













| General Information: |
|
| Name(s) found: |
pgl-3 /
CE17420
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 693 amino acids |
Gene Ontology: |
|
| Cellular Component: |
P granule
[IDA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP |
| Molecular Function: |
protein binding
[IPI]
RNA binding [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEANKRQIVE VDGIKSYFFP HLAHYLASND ELLVNNIAQA NKLAAFVLGA TDKRPSNEEI 60
61 AEMILPNDSS AYVLAAGMDV CLILGDDFRP KFDSGAEKLS QLGQAHDLAP IIDDEKKISM 120
121 LARKTKLKKS NDAKILQVLL KVLGAEEAEE KFVELSELSS ALDLDFDVYV LAKLLGFASE 180
181 ELQEEIEIIR DNVTDAFEAC KPLLKKLMIE GPKIDSVDPF TQLLLTPQEE SIEKAVSHIV 240
241 ARFEEASAVE DDESLVLKSQ LGYQLIFLVV RSLADGKRDA SRTIQSLMPS SVRAEVFPGL 300
301 QRSVFKSAVF LASHIIQVFL GSMKSFEDWA FVGLAEDLES TWRRRAIAEL LKKFRISVLE 360
361 QCFSQPIPLL PQSELNNETV IENVNNALQF ALWITEFYGS ESEKKSLNQL QFLSPKSKNL 420
421 LVDSFKKFAQ GLDSKDHVNR IIESLEKSSS SEPSATAKQT TTSNGPTTVS TAAQVVTVEK 480
481 MPFSRQTIPC EGTDLANVLN SAKIIGESVT VAAHDVIPEK LNAEKNDNTP STASPVQFSS 540
541 DGWDSPTKSV ALPPKISTLE EEQEEDTTIT KVSPQPQERT GTAWGSGDAT PVPLATPVNE 600
601 YKVSGFGAAP VASGFGQFAS SNGTSGRGSY GGGRGGDRGG RGAYGGDRGR GGSGDGSRGY 660
661 RGGDRGGRGS YGEGSRGYQG GRAGFFGGSR GGS |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
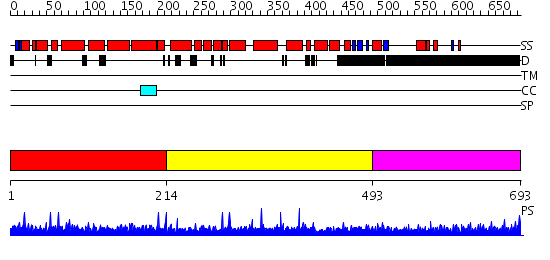
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..213] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [214..492] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [493..693] | 16.69897 | No description for 1y0fB was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)