













| General Information: |
|
| Name(s) found: |
cha-1 /
CE17308
[WormBase]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 627 amino acids |
Gene Ontology: |
|
| Cellular Component: |
synapse
[IDA neuron projection [IDA |
| Biological Process: |
acetylcholine biosynthetic process
[IMP]
|
| Molecular Function: |
acyltransferase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKEKVDELP PNDNWYETAL PKPPVPSLEA TLDRYLEYAA VVAVGQKASL ATTHDAAHKF 60
61 VRQATPLQEQ LLEIAEKSPN WATKFWLPEM YMRVRMPTPV NSNPGYIFPK VKFETKEDHI 120
121 KYTALLTRGL LEYKNLIDTK QVCREKSTGA QKLQMCMEQY DRVLSCYREP GVGEDTQIRK 180
181 QKTNDGNEHV LVMCRNQTFL LHSRINGALV SYADVEYQLA QIEEISKINQ NNTANIGASG 240
241 VGPRDNAALF WQDMLTVEQN SKSYEWVKSA LFVVCLDMED PIDYGKNDTM SISEKEKEFV 300
301 ARGYSTLTGH GSSKFGLNRW YDATIQLVVS SSGVNGLCIE HSTAEGIVII NMAETAIRYA 360
361 QKYFKSKMVW NDVRNVHPKS LTWHFSENSR NILKKQAEVF DELANELELE VLIFNEFGKD 420
421 SIKNWRVSPD GFIQLIMQLA HYKTHGHLVS TYESASVRRF GAGRVDNIRA NTQEALEWVT 480
481 AMASKKESKE RKLELFKKAV LKQVKVTLEN ISGYGVDNHL CALFCLARER EETTGEDIPS 540
541 LFLDPLWSEV MRFPLSTSQV TTSLDIPDCY LTYGAVVRDG YGCPYNIQPD RVIFAPTAFR 600
601 SDPRTDLQHF KKSLAGAMRD VKELLSN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
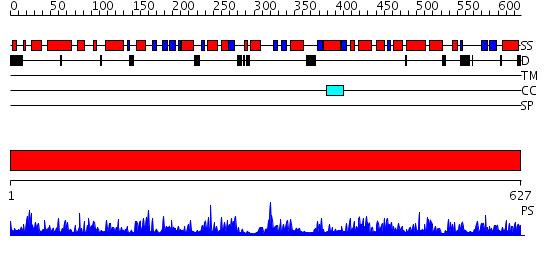
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..627] | 1000.0 | Structural Insights and Functional Implications of Choline Acetyltransferase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)