













| General Information: |
|
| Name(s) found: |
CE17112
[WormBase]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 786 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
proteolysis
[IEA reproduction [IMP |
| Molecular Function: |
zinc ion binding
[IEA subtilase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPKPPGPVAK RLKLKDDSVS GSDSASTRTS RKTRGKMMRS NEKIGTLTMD EAIKAAEKIT 60
61 GFQKIDLRLT HRRDSDAANS MKWISTFDED DVVIQRKLCM ICKQSPMDAF FKSHVAQAHP 120
121 ATPSEASEVM RRLSDVRYSR FCQNGQISKE SIQKMFPTIS PILLINFLAG LGFIICRKDY 180
181 NKKSVNFEKK YQGSDVRGVS KSEANEILNS LKQNSIRFLD ELEFSAEIVT NARMLDPSTQ 240
241 STVQCHICDK ISNLKNFTRH MQIHRDSLEI LGIKYSLDDV IEIFKRITDV RNTLYYAAQP 300
301 ISQSKIMEIC EGNVEVQRKL VSFLLDAGFV VADTSKYESL AQKIIVMDPE MPELIEVKEE 360
361 PEDCDEKYEL PILEVSEPGS SEPSESRTRS RSSSFPFSAF QQIVFYKNVS DTSSISSTSS 420
421 TESRIILRPC SSPSDEQFPY CVLNAIPEHF EVEHALFDIL EAMTDDSEFK LEKSQFGFLL 480
481 GYTMYCLVVA NGSKQNVISR LTLGDFRNSK IDEQSGLHYF SFSTKTTCKK LDTLEYLCAD 540
541 ERIWRALEIF EVARERRALE MKWSTLNENS APFFFSFVSP SPIVKDTNYW LSIFLRSCGI 600
601 DWLCKSNNIC SAAWSLVFRD SNKTSHQQYA ITYFKLLREK ERSQIQFLDG GLAGHPTYRK 660
661 ISSVVISKKR KNHIVPHEKI RKKQKRYRTP ERIGIPNRPR GGDHKSLDFK RRTGRYVEPE 720
721 SPEDSDETDK VQESVICHVK EDPIDSTYFD SCECIPTPLE MTSHSSSQLY QQYRVKVEGD 780
781 VTLTCL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
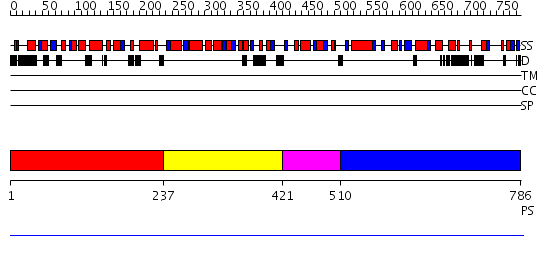
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..236] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [237..420] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [421..509] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [510..786] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)