













| General Information: |
|
| Name(s) found: |
CE16635
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 645 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotion
[IMP]
|
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEFIFNDDLK RQMDFLIDST EHATERIKLC PLLKKFSERF GLNYSTYLSR FHQKLCPQIS 60
61 NMDTYSVESR IRLLFALSGK VSDEFLARMQ SLGNVELYEN RRIEKFVSND ESLILEDNQY 120
121 GQLNDEKCKK FMEFLMQHTK DFVKPMSATQ VFEEFKKREN SSTHSSMYFE RFRNELAPMI 180
181 HLWNAYSVGA RVRLMFVLSQ SVENEFLTRI EQLGIVQLDD KKRIWKFTSF DEQLKLQGRH 240
241 GRCSVDIESP EFVRFVQFLV NKTEKSNEPI DQRRVFGEFR ALEHGVLDVT TYRGRFHGVV 300
301 ARNMILLDNL SIDLRIRMMF ALSGKMEANF LSEVEKQGIV RLDDQNRIIE YVANDGKMKL 360
361 KGQHEGRKRA WKTRATGANS GNTGSSGNYS KRSRRDGEGC SSRQETPHMD TAGSFDDSFN 420
421 FEEFTRMRLS YNDVLKSFAT VIDRAPKVKI SDDTKGNVDK HKAQQIIDLL KTFTTVVESA 480
481 PKLEIVDESE VQCDAVNNNK AQEADVEGSE YPKISMLNVL DKLDNMLISL ESELLNELLN 540
541 EIEQLRVGPA DKMIPLKLFI NAMESMLQNI TGKFQTKDAD GSTPAKDFLK TLTHFLKWMK 600
601 VPLLASFLKK VEEEIEKLGD IEQNVMIGDV NQQVRDLLVF LVVSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
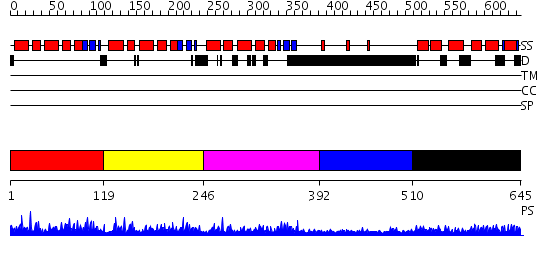
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 52.031517 | No description for PF04435.9 was found. No confident structure predictions are available. | |
| 2 | View Details | [119..245] | 59.508638 | No description for PF04435.9 was found. No confident structure predictions are available. | |
| 3 | View Details | [246..391] | 52.080922 | No description for PF04435.9 was found. No confident structure predictions are available. | |
| 4 | View Details | [392..509] | 2.065996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [510..645] | 2.055991 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)