













| General Information: |
|
| Name(s) found: |
nurf-1 /
CE15910
[WormBase]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 510 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proteinaceous extracellular matrix
[IEA |
| Biological Process: |
positive regulation of growth rate
[IMP cell adhesion [IEA gamete generation [IMP blood coagulation [IEA |
| Molecular Function: |
zinc ion binding
[IEA protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLRTVILPT ATYTAPSQYC PSTVLSNLTL SCPYQDQEAD ASLEISDFAE VLPKFESAEQ 60
61 DWNSFGYLLN EQPGTSSDKT TTPIKKITVF QKPVEPIGKG PRRRRRCADR EISELAAKPK 120
121 AEVKKEVINP ADITLGGDTY DYVKEQKPTE SIATNVSRRR RTSANLSKSE DDRDKPESQS 180
181 TAPKSKERRT SEPPASHVAF HTPGSATPHD INLSIEHCTC QKIFDASKLY IQCELCARWY 240
241 HGDCVGVAEQ TILGLEHWSC EECIEEQERV KDQPALYCVC QKPYDDTKFY VGCDSCQGWF 300
301 HPECVGTTRA EAEQAADYNC PACTREAEGY ESEASDVSGS SRVSVQLTRA DYTHVFELLE 360
361 LLLEHRMSTP FRNPVDLNEF PDYEKFIKKP MDLSTITKKV ERTEYLYLSQ FVNDVNQMFE 420
421 NAKTYNPKGN AVFKCAETMQ EVFDKKLIDV REQMTARQQM LLLATAQQQD PMSSIRKRVQ 480
481 SESQRTVDSL DIDSDQLLPL DANLMRLYDF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
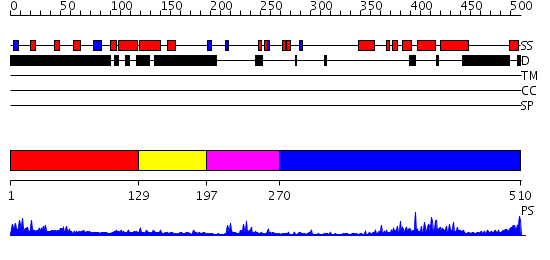
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..128] | 1.216995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [129..196] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [197..269] | 8.30103 | Solution structure of PHD domain in PHF8 | |
| 4 | View Details | [270..510] | 27.522879 | No description for 2f6jA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 |
|
||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.76 |
Source: Reynolds et al. (2008)