













| General Information: |
|
| Name(s) found: |
math-49 /
CE15279
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 423 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotion
[IMP]
nematode larval development [IMP] positive regulation of growth rate [IMP] embryonic development ending in birth or egg hatching [IMP] molting cycle, collagen and cuticulin-based cuticle [IMP] reproduction [IMP] growth [IMP] |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSRSFLLTH TFKDVSKLDE GGSYYSDIEI RHNIPWRLSI SKKDGFLGVN LHCENKECSE 60
61 TKKWSFQTKF TMKLVAVSGK FFQRTVQHEF QKPEGHGMDK LISWENMLRD YVDEDSIIIE 120
121 IHADIVSSSG FFEQKCLPNM PADSGKSFVL KHTFKNLSRI LEGEYQYSDV EVHYNIPWKI 180
181 ELQEENGSFG FHLHHVSEDR TKTVVVEREI KLIAANGKFH SVPKRTCTFG AKSGSVFLIY 240
241 CNGIEKDYVS NDSIDVEVRV KIIEVKDVPC LEKSFVLSHT IKNMSNIKEG GSIFTDIQTW 300
301 FNIPWRMKIE KQNGFLGLYL LHGKSQPESK KSSIEHDFQL KIVSPNGKIL SIERGGGVGW 360
361 DMFLRWDVLE EDYLVNDTII VEARVWIRKM TGMEDVEKFP GFVIPVGDQE YSLEKWLEMA 420
421 FDH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
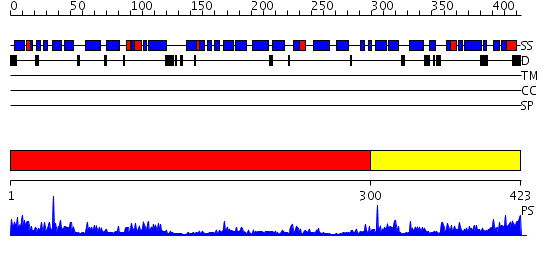
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..299] | 19.522879 | Crystal structure of HAUSP | |
| 2 | View Details | [300..423] | 3.64 | Crystal structure of the TRAF-like domain of HAUSP/USP7 bound to a p53 peptide |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)