













| General Information: |
|
| Name(s) found: |
sulp-4 /
CE12208
[WormBase]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 749 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA integral to membrane [IEA apical plasma membrane [IDA |
| Biological Process: |
sulfate transport
[IEA transport [IEA |
| Molecular Function: |
secondary active sulfate transmembrane transporter activity
[IEA oxalate transmembrane transporter activity [IDA chloride transmembrane transporter activity [IDA transporter activity [IEA sulfate transmembrane transporter activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPPGRINHA RHLSEAEEEA ENYLEHFYNK SEPNKRRVSF VQRGAMNQAQ FDEKFDYNKP 60
61 HLENELKKQA KKFVRRFYEP FTSFFALKLF IFDLIPILKW FPEYKWKTDL SLDIIGGITV 120
121 GVMQVPQGIA YALLAKQPAI NGLYTSLFPP LIYMLFGTSR HASLGTFAVV SLMTGLSVEK 180
181 LAAPTDYDPS SFNETDIDLV KLPSPTEVSC AITITMGLIL FVMGVLRLQF LTTYLSDQVI 240
241 AGFTVGSSVH VLVSQLKTLL GIRGLPRHSG PFYLFRHLYD LVMSITRANP VSCGISIVSI 300
301 IVLHFGKEFI NPFIKRKTKS NIPIPWELVI VILSTVFVAV TGVDTEAKVQ VVNKIPVGVP 360
361 NFSLPSFYLI PQVLPDAISI TVVSISVWLS ISKMLAKRYN YELDSGQELF ALSFASISSS 420
421 FIPSIPNSCS LSRTLVAVGA GCTTQLSIFF SSLIVFSVVI FLGTLLETLP MAALSAIICV 480
481 ALQGMFRKFA DLIDLWKVSK IDFTIWVVSC VSTILLDVST GLIISVCFAL FTTILREQYP 540
541 KWHLLASVKG TQDFRDAERY GETVYFKGIC IFRFDAPLLF HNVECFKKSI EKAYTEWQKS 600
601 HEFYVLREER ETILNTKLDG SDESIDGKMF QTAQSTLNTH SPDILSRHFV IDCSGFTFID 660
661 LMGVSALKEI FSDMRKRGIL VYFANAKAPV REMFEKCHFF NFVSKENFYP TMRDATSIAR 720
721 QRQLELGFKD TGYVPEHDRL TEVLSSHPM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
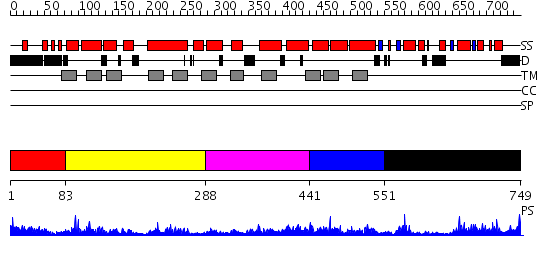
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 1.233989 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [83..287] | 6.946992 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [288..440] | 1.958986 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [441..550] | 3.950996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [551..749] | 2.46 | SOLUTION STRUCTURE OF SPOIIAA, A PHOSPHORYLATABLE COMPONENT OF THE SYSTEM THAT REGULATES TRANSCRIPTION FACTOR SIGMA-F OF BACILLUS SUBTILIS, NMR, 24 STRUCTURES |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 |
|
| Source: Reynolds et al. 2008. Manuscript submitted |

|