













| General Information: |
|
| Name(s) found: |
ldb-1 /
CE11352
[WormBase]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 578 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
mechanosensory behavior
[IMP vulval development [IMP locomotory behavior [IMP hermaphrodite genitalia development [IMP locomotion [IMP multicellular organismal development [IEA positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP |
| Molecular Function: |
transcription factor activity
[IDA protein binding [IDA transcription cofactor activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFHRPPSARP PLNHVDQLKS LQATALGKPG MRSQATEPQP IGNTVSPLEF RIHDMNRRLY 60
61 IFSSTGVSEN DQQQWWDAFS HEFFDDDCKL WFVIGSEPVA FASRERYIIN RQFIPKFFRS 120
121 IFDSGMRELQ YVLRGPSREC TLANGSQAYE NENVLQITRY DQSSQFEVNT EGKLYVEFAP 180
181 FDEVMNYRIK AWTLELKRSN EFVYNQNTAD YRVEAQNPEQ ENKPRMGFFK STFNLMTMLK 240
241 ILDPMQSIMS SAKSAPAITP REVMKRTLFQ HHQVRQQNMR QQQLNQQMMI PAPEPEKPKP 300
301 ARKRQRKPAA NPRGSKKATA AAAAAAAAAT NGVPPTVPTA SPANNQQFPP NPMTSQFQQM 360
361 SYPDVMVVGE PSMMGSEFGE NDERTISRVE NSQYDPNAMQ MQSLGQGPNS SMNINGRNMM 420
421 NQHHPGMQPP PGQQHMPPHS MGSQMPTSMH NPMHMPPGSM QGHGGMPPMP STMANQMPPP 480
481 NLPPHTMSNQ MPNSRMPPMQ GQMPPSGMPG QMSNPNMMGS GMPPMSMSSQ MPGSMSNPMP 540
541 NQMPGGMQMN QMPPPNYSQY TGGPPPQWPP PNSAMITG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
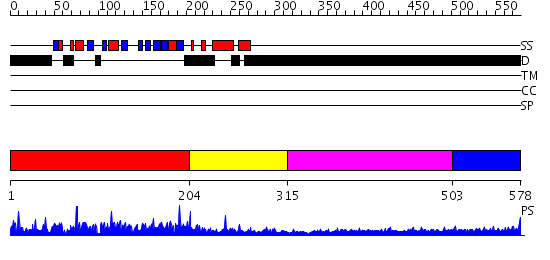
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..203] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [204..314] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [315..502] | 6.69897 | Sec24 | |
| 4 | View Details | [503..578] | 1.14 | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)