













| General Information: |
|
| Name(s) found: |
ldb-1 /
CE11350
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 592 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IEA |
| Biological Process: |
mechanosensory behavior
[IMP vulval development [IMP locomotory behavior [IMP hermaphrodite genitalia development [IMP locomotion [IMP multicellular organismal development [IEA positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP |
| Molecular Function: |
transcription factor activity
[IDA protein binding [IDA transcription cofactor activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYGHHPDYGY GNYGHPPPFA GPESSNSHYG MPPSQGTNSQ NNMMMRSQAT EPQPIGNTVS 60
61 PLEFRIHDMN RRLYIFSSTG VSENDQQQWW DAFSHEFFDD DCKLWFVIGS EPVAFASRER 120
121 YIINRQFIPK FFRSIFDSGM RELQYVLRGP SRECTLANGS QAYENENVLQ ITRYDQSSQF 180
181 EVNTEGKLYV EFAPFDEVMN YRIKAWTLEL KRSNEFVYNQ NTADYRVEAQ NPEQENKPRM 240
241 GFFKSTFNLM TMLKILDPMQ SIMSSAKSAP AITPREVMKR TLFQHHQVRQ QNMRQQQLNQ 300
301 QMMIPAPEPE KPKPARKRQR KPAANPRGSK KATAAAAAAA AAATNGVPPT VPTASPANNQ 360
361 QFPPNPMTSQ FQQMSYPDVM VVGEPSMMGS EFGENDERTI SRVENSQYDP NAMQMQSLGQ 420
421 GPNSSMNING RNMMNQHHPG MQPPPGQQHM PPHSMGSQMP TSMHNPMHMP PGSMQGHGGM 480
481 PPMPSTMANQ MPPPNLPPHT MSNQMPNSRM PPMQGQMPPS GMPGQMSNPN MMGSGMPPMS 540
541 MSSQMPGSMS NPMPNQMPGG MQMNQMPPPN YSQYTGGPPP QWPPPNSAMI TG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
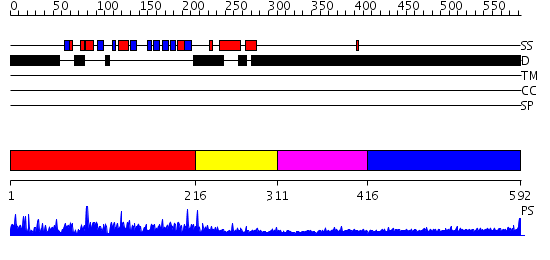
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [216..310] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [311..415] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [416..592] | 7.30103 | Sec24 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)