













| General Information: |
|
| Name(s) found: |
stau-1 /
CE11107
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA |
| Biological Process: |
rRNA catabolic process
[IEA |
| Molecular Function: |
RNA binding
[IEA ribonuclease III activity [IEA double-stranded RNA binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQAVFETTLT QKMDGVMIVQ ETTTDLADTL ENASISAEKS EQKPERLHPQ HWCGQHKFEA 60
61 DSPTNFYDYT NAKEKEKSAM CRVAEIARFN KLRHVYNLQD ESGPAHKKLF TVKLVLTEAE 120
121 TFEGSGTSIK RAQQASAEAA LKGTKLPLPT EKPTKKRIND TTKPHRVLQN VCRTLQYQMP 180
181 NYISCNPPVY PDPGCPLPEH ILLPLESMAL YAPPFPTLPI DPARPQGPKL QAVIVNINGK 240
241 SIATGIGETY PLAKQDAAAK ALAVLSPLLR EHQNGSDNGF GKENIPVHKQ KSVISDIHEK 300
301 AYQLKVNVVF EVLKEEGPPH DRQYVVRCAF VTSGNVVKAE AVGKGKKKKS AQQEACTQLL 360
361 ATVEHLTPEN NPVALATNVC KTQKKLAAMN REPKRKTIVK DKKMDPLYGH QINPVSRLIQ 420
421 VTQAKSKEHP TFELVAEHGV SKYKEFIIQV KYGDDVQEGK GPNKRLAKRA AAEAMLESIG 480
481 FVKPLPPPGK SLLKKMIDCD PSLPEISHWT GPPPTAVSVS TSEPDTSEAA QLSPEQTDIS 540
541 EKRELSPDTE KRRVTFNSQV HACPPPGDQD YPNSIVQSLK KDAIVEGKIR RLKRSKENRR 600
601 ALTAEQIVEL SERAQSYLQT KNTTIQSSQS SSAHHHLEQL SDFFKFSLQY TSFPQVGIDQ 660
661 HFTIVSIGLE APLVGHGTGC STTEADENAA LDAIAKLKEL SASKT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
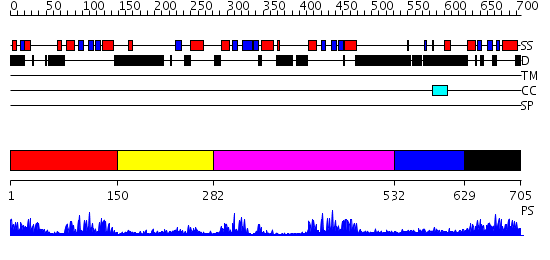
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..149] | 35.0 | RNase III endonuclease catalytic domain; RNase III, C-terminal domain | |
| 2 | View Details | [150..281] | 1.14 | Solution structure of the dsRBD from hypothetical protein BAB26260 | |
| 3 | View Details | [282..531] | 22.69897 | No description for 2yt4A was found. | |
| 4 | View Details | [532..628] | 7.0 | PKR kinase domain- eIF2alpha- AMP-PNP complex. | |
| 5 | View Details | [629..705] | 2.13 | Staufen, domain III |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)