













| General Information: |
|
| Name(s) found: |
mec-10 /
CE09444
[WormBase]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 724 amino acids |
Gene Ontology: |
|
| Cellular Component: |
membrane
[IEA integral to membrane [IEA |
| Biological Process: |
mechanosensory behavior
[IMP sodium ion transport [IEA ion transport [IEA response to mechanical stimulus [IMP |
| Molecular Function: |
ion channel activity
[IEA sodium channel activity [IEA amiloride-sensitive sodium channel activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNRNPRMSKF QPNPRSRSRF QDETDLRSLR SFKTDFSNYL ASDTNFLNVA EIMTSYAYGE 60
61 SNNAHEKEIQ CDLLTENGGI EIDPTRLSYR ERIRWHLQQF CYKTSSHGIP MLGQAPNSLY 120
121 RAAWVFLLLI CAIQFINQAV AVIQKYQKMD KITDIQLKFD TAPFPAITLC NLNPYKDSVI 180
181 RSHDSISKIL GVFKSVMKKA GDSSSEALEE EEETEYDMNG ITIQAKRKKR GAGEKGTFEP 240
241 ANSACECDEE DGSNECEERS TEKPSGDNDM CICAFDRQTN DAWPCHRKEQ WTNTTCQTCD 300
301 EHYLCSKKAK KGTKRSELKK EPCICESKGL FCIKHEHAAM VLNLWEYFGD SEDFSEISTE 360
361 EREALGFGNM TDEVAIVTKA KENIIFAMSA LSEEQRILMS QAKHNLIHKC SFNGKPCDID 420
421 QDFELVADPT FGNCFVFNHD REIFKSSVRA GPQYGLRVML FVNASDYLPT SEAVGIRLTI 480
481 HDKDDFPFPD TFGYSAPTGY ISSFGMRMKK MSRLPAPYGD CVEDGATSNY IYKGYAYSTE 540
541 GCYRTCFQEL IIDRCGCSDP RFPSIGGVQP CQVFNKNHRE CLEKHTHQIG EIHGSFKCRC 600
601 QQPCNQTIYT TSYSEAIWPS QALNISLGQC EKEAEECNEE YKENAAMLEV FYEALNFEVL 660
661 SESEAYGIVK MMADFGGHLG LWSGVSVMTC CEFVCLAFEL IYMAIAHHIN QQRIRRRENA 720
721 ANEY |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
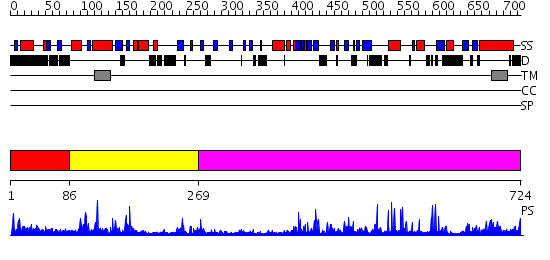
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..85] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [86..268] | 3.8 | No description for 2qtsA was found. | |
| 3 | View Details | [269..724] | 82.69897 | No description for 2qtsA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.24 |
Source: Reynolds et al. (2008)