













| General Information: |
|
| Name(s) found: |
uri-1 /
CE09016
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 446 amino acids |
Gene Ontology: |
|
| Cellular Component: |
prefoldin complex
[IEA |
| Biological Process: |
hermaphrodite genitalia development
[IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP positive regulation of cell proliferation [IGI germ-line stem cell division [IMP DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis [IGI gamete generation [IMP protein folding [IEA positive regulation of mitotic cell cycle [IGI reproduction [IMP DNA damage checkpoint [IGI locomotory behavior [IMP morphogenesis of an epithelium [IMP positive regulation of meiotic cell cycle [IGI |
| Molecular Function: |
unfolded protein binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSELYVAECN AAKARLEVET ECRRIIADNY EKVLERAKEY SKKLEAPILA KICEVGYFCG 60
61 AHVVRTNQVT MEMGANYYGE CSIHTAEKIL DRRVSEMRRQ REEGLDSIRM LDDKIKFAQD 120
121 NFSSVSIDGG PIEIVEKYDE EMEKLFYQNR KNKKMSQNDE QNEKIIEKNK LNGDQDHNDV 180
181 MNRLEELEKL EQDNDEMEEA PIPKPFEVSN EDLQDLGKLE EVEEVEQDPT DKKLRFGMNE 240
241 RLLAQPGVTQ KEMERLLDFL DTCDEESSDE EYDEDEEEGD DDEENLNNSV EELDSDDYAS 300
301 DDNIEKSNNS DQQLKSKKIE TQVIKVEEIV EETVTKTTVK RKKSGVRFAQ QLENVKTFHQ 360
361 TDTVEIKEED SVIETKSILR STEPTPVDRS ALEPEQKELA AMSTMTFPGE IVEKNPYECE 420
421 PSTSRDPAPV ITEKKVSKFR ASRHRN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
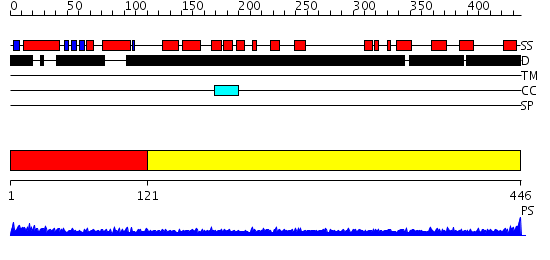
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 3.63 | Prefoldin alpha subunit | |
| 2 | View Details | [121..446] | 3.09691 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)