













| General Information: |
|
| Name(s) found: |
nab-1 /
CE08684
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 721 amino acids |
Gene Ontology: |
|
| Cellular Component: |
presynaptic active zone
[IDA]
synapse part [IDA] plasma membrane [IDA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
protein binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTASELLSD DARARFSHTK ALFEQLERQQ DVPSFYSPRL QRQPPPPLPP KPPSQCPPSP 60
61 MSQVERNFSD LAADLDRIQS SPATSRFIQN KSSTLPSSYS DSTQQYSFEN TVATSQYGGL 120
121 QTNNNNNNIN MNSFEPYWRN GSIYRRQFEG NGKPFGEEND GISPTTNGVK HTTYAVVKTP 180
181 KETELETTSE NRGLMETRRG LSPERPDVDL TNRKVSFSTA PIPVFSAFSV EDYDRKNEDI 240
241 DPVASCAEYE LERRLERMDL FEVDLEKGAE GLGVSIIGMG VGADSGLEKL GIFVKSITPG 300
301 GAVHRDGRIR VCDQIVSVDG KSLVGVSQLY AANTLRSTSN RVTFTIGREQ NLEESEVAQL 360
361 IQQSLEHDRM RMMGDEEDDI EEPPPPPSQM PQLPIEQDRP TTSMITKEEI EIRSKIAALE 420
421 LELDVTHKKA EQYHEVLSST KSHCDQLEKQ NEQANYMIKN YQEREKELLN REENHVEQLR 480
481 DKDVHYASLV RQLKERIDEL ESKLEEAEER RHSIQNLELI ELREKLKEKV EKRNEGMAYR 540
541 PGGELPHEDK AVMVNLETIT PSTKKDAEVS VGSSWTEEYS SPCESPVPRI SEPASPALPH 600
601 KLTHRKLLFP LRKKYAENEF WRATCQPVGL QALHWTVDDV CQLLVSMGLD KYVPEFTINK 660
661 IDGAKFLELD GNKLKAMGIQ NHSDRSLIKK KVKGMKNKIE RERKQLERES RTRVIAHTIP 720
721 M |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
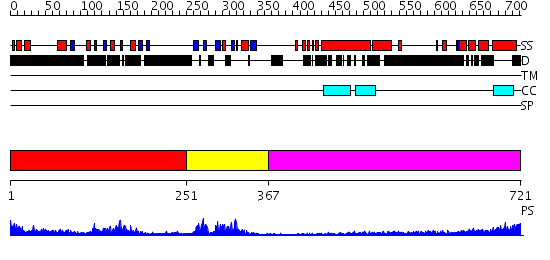
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..250] | 19.39794 | solution structure of the split PH-PDZ Supramodule of alpha-Syntrophin | |
| 2 | View Details | [251..366] | 23.39794 | Solution structure of the PDZ domain of Spinophilin/NeurabinII protein | |
| 3 | View Details | [367..721] | 31.154902 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)