













| General Information: |
|
| Name(s) found: |
erm-1 /
CE07806
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 564 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoskeleton
[IEA cytoplasm [IEA cell cortex [IDA filamentous actin [IDA membrane-enclosed lumen [IDA extrinsic to membrane [IEA |
| Biological Process: |
locomotory behavior
[IMP morphogenesis of an epithelium [IMP nematode larval development [IMP positive regulation of growth rate [IMP embryonic development ending in birth or egg hatching [IMP protein amino acid dephosphorylation [IEA actin filament organization [IMP gamete generation [IMP tube formation [IMP growth [IMP reproduction [IMP positive regulation of multicellular organism growth [IMP |
| Molecular Function: |
protein tyrosine phosphatase activity
[IEA structural molecule activity [IDA cytoskeletal protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVAAGDINVR VTSMDSELEF AIQSSTTGKQ LFDQVVKTIG LREIWYFGLQ YTDNKGFPTW 60
61 LKLNKKVLSQ DVKKDPTLVF KFRAKFYPED VAEEIIQDVT MRLFYLQVKD GILSDEIYCP 120
121 PETSVLLASY AMQAKYGDYV PETHVAGCLT ADRLLPQRVL GQFKLNSEEW ERRIMTWWAD 180
181 HRATTREQAM LEYLKIAQDL EMYGVNYFEI RNKKGTDLYL GVDALGLNIY DKADRLSPKV 240
241 GFPWSEIRNI SFNDKKFVIK PIDKKAHDFV FYAPRLRINK RILALCMGNH ELYMRRRKPD 300
301 TIEVQQMKQQ AREDRALKIA EQEKLTREMS AREEAEQRQR DAEKRMAQMQ EDMERARLEL 360
361 AEAHNTIHSL EAQLKQLQLA KQALEQKEYE LRELTAQLQS EKAMSDGERR HLRDQVDARE 420
421 REVFSMREEV ERQTTVTRQL QTQIHSQQHT QHYSNSHHVS NGHAHDETAT DDEDNGATEL 480
481 TNDADQNVPQ HELERVTAAE KNIQIKNKLD MLTRELDSVK DQNAVTDYDV LHMENKKAGR 540
541 DKYKTLRQIR GGNTKRRIDQ YENM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
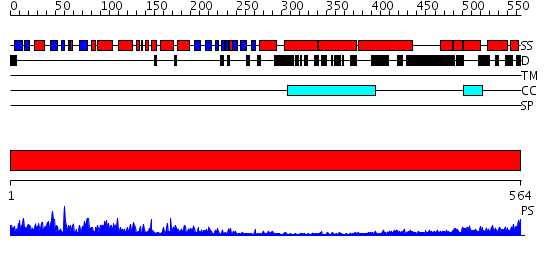
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..564] | 89.30103 | No description for 2i1jA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)