













| General Information: |
|
| Name(s) found: |
wapl-1 /
CE07433
[WormBase]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 746 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
locomotory behavior
[IMP embryonic development ending in birth or egg hatching [IMP positive regulation of growth rate [IMP oviposition [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSDANSDDP FSKPIVRKRF QATLAQQGIE DDQLPSVRSS DSPDVPDTPD VPVNQLSSPP 60
61 LSLPETLSEG NAETNLSDDS EPEMLSQSST SSLNRRMEDS AIDPSRGTRK SQSRGFDYDP 120
121 AGERTTAPVQ KKKKDEIDMG GAKFFPKQEK KHVYTHKWTT EEDDEDEKTI SSSSNRYSSR 180
181 PNQPAVSARP RQPVYATTST YSKPLASGYG SRVRHIKEAN ELRESGEYDD FKQDLVYILS 240
241 SLQSSDASMK VKCLSAISLA KKCVSPDFRQ FIKSENMTKS IVKALMDSPE DDLFALAAST 300
301 VLYLLTRDFN SIKIDFPSLR LVSQLLRIEK FEQRPEDKDK VVNMVWEVFN SYIEKQEVGG 360
361 QKVSFDMRKE SLTPSSLIIE ALVFICSRSV NDDNLKSELL NLGILQFVVA KIETNVNLIA 420
421 DNADDTYSIL ILNRCFRILE SSSVFHKKNQ AFLISHRSNI LISSLAKFLQ VILDRVHQLA 480
481 EEEVKKYISC LALMCRLLIN ISHDNELCCS KLGQIEGFLP NAITTFTYLA PKFGKENSYD 540
541 INVMMTSLLT NLVERCNANR KVLIAQTVKM VIPGHDVEEV PALEAITRLF VYHESQAQIV 600
601 DADLDRELAF DEGGCGDEEE EEEGGDESSD EDGVRKDGRL DRNKMDRMDQ VDVVHALQQV 660
661 MNKASAHMEG SVIASYHALL VGFVLQQNED HLDEVRKHLP GKNFQNMISQ LKRLYDFTKA 720
721 TMAKRVESNS GFRAIERVIE YLERLE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
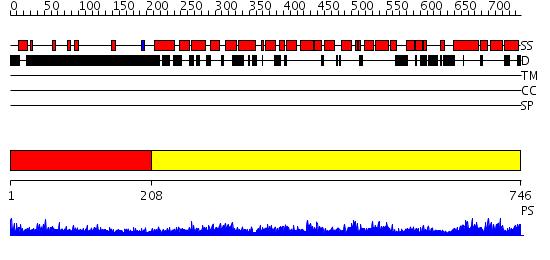
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..207] | 13.233995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [208..746] | 1.04 | No description for 2gl7A was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)