













| General Information: |
|
| Name(s) found: |
let-99 /
CE06147
[WormBase]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 698 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell cortex
[IDA |
| Biological Process: |
pronuclear migration
[IMP embryonic development ending in birth or egg hatching [IMP gamete generation [IMP |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSADYSSEKF KATHLFDEIL WHFRSNLSLK TNRRGLATAE NSFSGKEAVD FLMIEMPRII 60
61 PNNVPERDKM QKFLEFMMDM NVISEAFPKK VKQRRPFSES RIYLFMKTLD ELKHPKPRSR 120
121 RSASFSGARK SAKVAQPASP AATMHRPPKA RLPRRLSRSN GNIDKAGIDN SSGGVENHGF 180
181 DDHKDDEIPK KQRTPKILNR SLESICTEEY TEKREVSEMK EKVYDWLPFF KSRRNHTKVN 240
241 QPTRRSASLD RNHCVLEQEK AEAALRSQKV TTPPIREAPK DVVFMHPQGP LPAVPSRYQN 300
301 HRTSIAGSNP ALLSRGRMYE SIMRRSSVVP VADSVQANNE CSFWKTELLG RLEQIYDRTL 360
361 PCEWASKVDG YDIQWNMIEI DHADGIVKSR CQGLQPDYPQ TVIQFMDYLV RYPFVTHKKM 420
421 DTGLEYNVNR IFITLVNRLE DLNAPLQFDE CSLIVNLLCK IDSFAAMLDN GPARRWSKVM 480
481 ISSSASSIEE AGLMVDGFSR DLPACGIRAS KYRRRALSPF DNRVNLEIQD EKSYEIREQW 540
541 LIEAIQLVLL SLPTSRRRKL HKFVTFIQSI ETNAVFDLAD PSNGSSNNRE AAIIGLWTGV 600
601 CSKCRKQQGM LITAVLLANF QSLFAVPVEF IEQVKRLECE EKDRYSGPRY AKITRSRNDW 660
661 QPPVPDKPQA SPAVFKKPLR EVACEKTKKG LFTRLLRK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
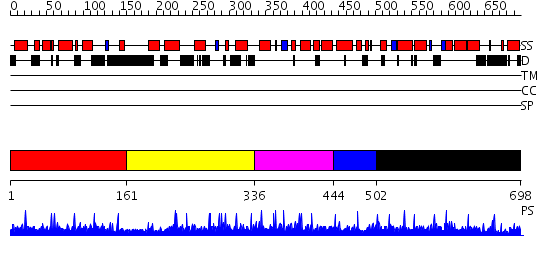
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..160] | 1.81 | No description for 2ysrA was found. | |
| 2 | View Details | [161..335] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [336..443] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [444..501] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [502..698] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)