













| General Information: |
|
| Name(s) found: |
CE03637
[WormBase]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 796 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
glycerol metabolic process
[IEA |
| Molecular Function: |
glycerophosphodiester phosphodiesterase activity
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MHFTRRAVSP RASFVFDRHV GTINSSLSRR PRISECVEEE EEDGGGFDLF EEMRQPIQEN 60
61 IPMIILEEEE DDNDNLVMSV ARPVRVHFAV DVENLHAHQS VYVVGSNDVL GTWEATRAMP 120
121 LVQDPDRFMR WKGSIVTDVH QLKFRYFIGY NLMSDQGERL IVDKWEAFLH PRSTLCLAES 180
181 RNDECRVDRV DLFGYYAGRK CVSDGWLQYP DENQILLRLH GNALKFYKTA KERKNCRVKM 240
241 TPLDVRFKAP PSGHISFSYG EDEEDEEEDQ NVPSNKCTHS ATHVAVLSDP RPKFYDQEDT 300
301 GVTFNNNKDY LVFRTHCLAA EFLAFYIEIF SEERKRIGAC YALPSSLQDA SGIIHLPFIN 360
361 SSGRPIGQVS IEYLCVRALT RLDGPQFMDQ TYCRHWKKRN ALEVGHRGAG NSYTKFAMAR 420
421 ENTIHSLNTA AKNGADYVEF DVQLTKDRIA VIYHDFHVLV SVARRDGLAM PPPMTREQLD 480
481 SSNLDYHELP VKDLKLSQLK LLMLDHLSFP QKKENVKKLV EAGEEEEDFK PFPTLLEALT 540
541 KVDPDVGFNV EVKYPMMQNN GEHECDHYFE RNLFVDVILA DVMKHAGNRR IMFSSFDPDI 600
601 CSMVATKQNK YPVLFLCVGE TQRYTPFQDQ RTSTSMTAVN FAAGADLLGV NFNSEDLLKD 660
661 PMPVKKANEF GMVTFVWGED LDKKENINYF KKELGVDGVI YDRIGEEERR RNVFIVEREQ 720
721 KRALLSCSGA STPQRAASPS PVSSENNNTS PPHSAKLTRN AQSDSALLEE NEDDVEHLSD 780
781 KLNITGFQSA PMVSTK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
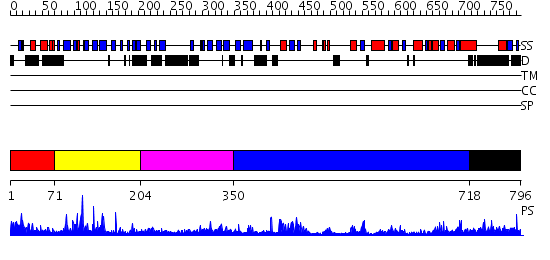
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | 2.080988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [71..203] | 33.522879 | CYCLODEXTRIN GLUCANOTRANSFERASE (E.C.2.4.1.19) (CGTASE) | |
| 3 | View Details | [204..349] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [350..717] | 53.39794 | Crystal structure of periplasmic glycerophosphodiester phosphodiesterase from Escherichia coli | |
| 5 | View Details | [718..796] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 |
|
||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)