













| General Information: |
|
| Name(s) found: |
CE03398
[WormBase]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 638 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
tRNA processing
[IEA embryonic development ending in birth or egg hatching [IMP |
| Molecular Function: |
FAD binding
[IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLSKSTRLLR RCFQTDVDVI VIGGGHAGCE SAAAAARCGS NTVLVTQNKN TIGEMSCNPS 60
61 FGGIGKGHLI REVDALDGLC ARICDKSAIT YQALNRAQGP AVLGLRAQID RKLYKTQMQN 120
121 EINSTKRLEI LEGEVAELLV ENGKIVGIRM MNETVIRTKC VVITTGTFLR AQIYQGMKTW 180
181 PAGRIGEKSS DRLSESFLKH GFELGRLRTG TPPRLMKDSI NFSKFERVAP DRTPIPFSFL 240
241 TKNVWISYED QLPTYLGHTN DEVCRIGNEN MHENYQVASE TTSPRYCPSL ESKLLRFPKL 300
301 HHRLFLEHEG LDSPHIYPQG MSLTFKPEVQ TQLLRAIPGL ENVEIFQPGY GVQYDFVNPK 360
361 QLKRTLETRK VEGMFLAGQI NGTTGYEEAA AQGVVAGINA SARAQNEPGM EVSRTEGYIG 420
421 VLIDDLTSLG TNEPYRMLTS RAEFRLYLRP DNADIRLTEL GRRHNAISDN RWAIFTETKG 480
481 ELNNLTQRTE EMKMSMVKWK RIIPKLAATS RNDGKVLSAF DLIHRYDLDK SDLELCLKDK 540
541 NIGEDILERL KIEGRYQMEH ERMKAKKQEI DRESATAIPD NTDFSTMRGM SLECIEKLER 600
601 ARPRNLAAAT RISGITPEAI VVLMRHLKNP APVRSSAV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
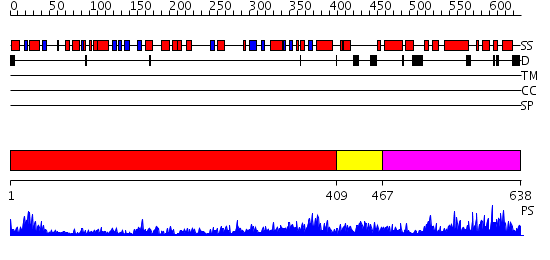
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..408] | 27.221849 | No description for 2gqfA was found. | |
| 2 | View Details | [409..466] | 2.239993 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [467..638] | 2.186968 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)