













| General Information: |
|
| Name(s) found: |
rpy-1 /
CE01807
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 596 amino acids |
Gene Ontology: |
|
| Cellular Component: |
synapse
[IEA |
| Biological Process: |
lipid storage
[IMP]
|
| Molecular Function: |
zinc ion binding
[IEA protein binding [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGQRQAKQHM QAGVKLYHQR HYAQAINKWR QSLNRLNNAE DRFITLGYLA QALCDQGEYE 60
61 GMLSYALSQM QLATDQNDSA MKCEAFLNLA KAYERLADFT KALQYGKASL EHPSMDPRTP 120
121 GYAHLTIALA HLGMSQFQQC LESFESAMNV ANETSDRLLE LQICVGLGSL FTLLRDITKA 180
181 LIFLRNALAI VQSVTVDDVH AKYRCLILYH LSVALRMKGS LVDAKEACDE ASQLAVEMGN 240
241 RAIHARCMCS LADIYRELGE SEAKETITKS WARYEDAYRV MRGANDKMGE VLVLSSMAKS 300
301 ASESRSHYTG QCECQAIQLN KKCIEIANQI GCKHVVLKCH LRLAELYSQL NDDDSEETAR 360
361 RAASRLTQEM QLFCNFCGQR YGLKDESLQA LRCSHIFHEK CLHTYLLQRT DQTCPKCRCR 420
421 AVLSDNISIR SSIASTIDVQ SPSTSFAPPG GVATPLVSAA AELGDITPQA TLTRRSQQDK 480
481 ILRGAEKDID RVDRSLARLK SQQSAAANAC SAVAVPSPEV PSTTASCASE PILTASTTSQ 540
541 SHTHSNANHK PPPPPRKPCC SQAALPPSSI VALPPAMPLA EDGPLVITHT PTVTDV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
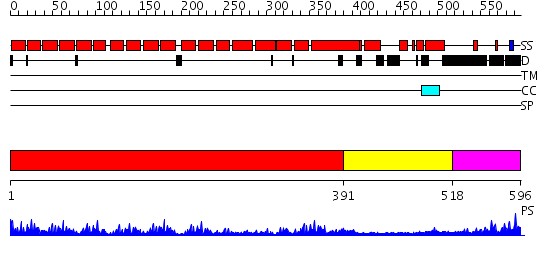
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..390] | 42.0 | The superhelical TPR domain of O-linked GlcNAc transferase reveals structural similarities to importin alpha. | |
| 2 | View Details | [391..517] | 3.09691 | Crystal Structure of the Hypothetical protein VPA1032 from Vibrio parahaemolyticus, Northeast Structural Genomics Target VpR44 | |
| 3 | View Details | [518..596] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)