













| General Information: |
|
| Name(s) found: |
set-2 /
CE01158
[WormBase]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 739 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
chromatin modification
[IEA |
| Molecular Function: |
nucleic acid binding
[IEA histone-lysine N-methyltransferase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYNNSAPYLN HSSLNTVRKK VVTVRRVLPS LPPPPPPPPS LYPPCSVFKV PYIPQRVYRS 60
61 INSTLYNNQS FASSSRGFYR KQKPIPKSHP KHQEHHHHAK ASVSTPVHSS STSRNSSVAP 120
121 TPQRTVSTSS SSSSAATSAR VSEDESDSDS TPGEVQRRKT SVLSNDKRRR RASFSSTSIQ 180
181 SSPERQRDVS SSSRTSSSSS TSSMKQEETA DEKSRKRKLI MSSDESSTTG STATSVVSSR 240
241 QSSLEPQQEK TDGEPPKKKS QTDFISERVS KIEGEERPLP EPVETSGPII GDSSYLPYKI 300
301 VHWEKAGIIE MNLPANSIRA HEYHPFTTEH CYFGIDDPRQ PKIQIFDHSP CKSEPGSEPL 360
361 KITPAPWGPI DNVAETGPLI YMDVVTAPKT VQKKQKPRKQ VFEKDPYEYY EPPPTKRPAP 420
421 PPRFKKTFKP RSEEEKKKII GDCEDLPDLE DQWYLRAALN EMQSEVKSAD ELPWKKMLTF 480
481 KEMLRSEDPL LRLNPIRSKK GLPDAFYEDE ELDGVIPVAA GCSRARPYEK MTMKQKRSLV 540
541 RRPDNESHPT AIFSERDETA IRHQHLASKD MRLLQRRLLT SLGDANNDFF KINQLKFRKK 600
601 MIKFARSRIH GWGLYAMESI APDEMIVEYI GQTIRSLVAE EREKAYERRG IGSSYLFRID 660
661 LHHVIDATKR GNFARFINHS CQPNCYAKVL TIEGEKRIVI YSRTIIKKGE EITYDYKFPI 720
721 EDDKIDCLCG AKTCRGYLN |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
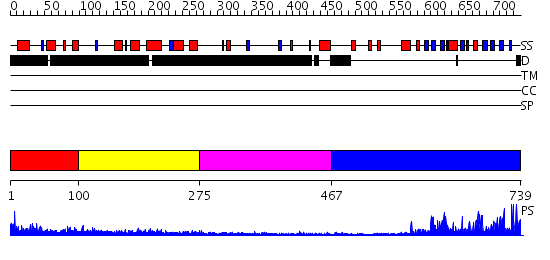
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..274] | 1.02 | Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition | |
| 3 | View Details | [275..466] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [467..739] | 56.522879 | No description for 2r3aA was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)