













| General Information: |
|
| Name(s) found: |
rsp1 /
SPBC11B10.05c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 494 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA interphase microtubule organizing center [IDA cytoplasm [IDA equatorial microtubule organizing center [IDA spindle pole body [IDA |
| Biological Process: |
selection of site for barrier septum formation
[IMP establishment of nucleus localization [IMP establishment or maintenance of cell polarity [IMP interphase microtubule organizing center assembly [IMP equatorial microtubule organizing center disassembly [IMP protein folding [RCA |
| Molecular Function: |
Hsp70 protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARTAFHDEF VDYYTILGAE STSSYVEIRQ QYLKLVLRYH PDRNPGREAE VLPQFQLIQK 60
61 AHEVLKDPKL RELFDQRRLL EAGRPDGVLR FRPKKSGPKN DISTKVASKV SVTMATKFAE 120
121 KKKKQDRENV DSKDNNITNF SLHRSFSASG KMEKNNSFKE VSTSKSYISS GYLHPKTSPI 180
181 FKKNGYATEN VVDPISSSPR FKGPNYNKFN AKLYLESLRE KRRTYTPLSE ISNGLNSNGV 240
241 ENSSITKSSP RSSSSSNNER FKDTSEESII FTSPNTPEHP SVYQTDITPE IKLEHSDNNS 300
301 PSKPEIPFRH PTSKPLPPKP LSRSKSSSLS RNQTRSQLND LSAENDSTSN STEYDDQLQS 360
361 ILRSLAIEGD DDEVAKVLPK PPSVPTIQAP IPPEAPRNLT NASVDSYLNS FEMYQRRWSS 420
421 YSIIYTQYAF QWQIFKNKCF QLDLMNTPGQ SRLIDNWKEG SQAIQLFYAY EQMHLRALEE 480
481 LQSLKESLFA SFGI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
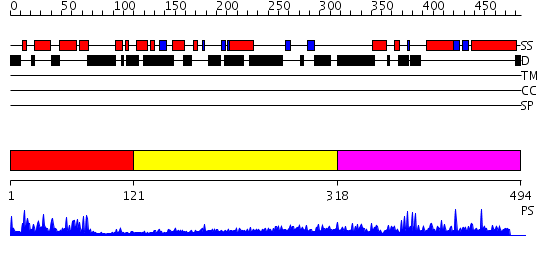
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | 27.0 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [121..317] | 1.714996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [318..494] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)