













| General Information: |
|
| Name(s) found: |
SPBC1734.07c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 618 amino acids |
Gene Ontology: |
|
| Cellular Component: |
TRAPP complex
[ISS nucleus [RCA cytosol [IDA |
| Biological Process: |
ER to Golgi vesicle-mediated transport
[ISS autophagy [ISS peroxisome degradation [ISS intracellular protein transport [ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MESNLSAKGI FQHASANASG ELFQEPVTED LEKVMQSKGL YSMEELCSII QEAYAPRVSV 60
61 LCSRDADEFA MRKGYPKGFW ELLYPFGDRI RGKVNRRGMN GEMLTLENLN LHFVPIDALE 120
121 SHLSEASFPS LRSVLCERYP GLVSPEPPSD YSFGVHYKNV ERWAWTLAHE GNYQMPLPVY 180
181 VRQLLTGIPI TPFETFSHPV AHLVVVTSHN PSPFESLRSL INSIPYLSLP AFVFNDINYL 240
241 FVYVHDEDQH DLELSMAIFD TMKQTFGDCG YFLRLHSQKA TLDYEHTVPF PTSSWLSAEE 300
301 RLHLLSNTDT EIRLLFESDN ESLRRLSSHI AFNGIVSYLD KCVRAWDDQY ASPRRGITGK 360
361 LLFASRKYLS SSNASTNSNY FPSSNAYRPF SPEAYLRKLA DYSFMLRDYS HANQIYEIAS 420
421 RQYENDGACL YSAASLEMIV ITEHILHLKM PYMSLTNTLR INEYMQSAML NYLNKSFNSY 480
481 YHAARCFLLL GQFLSCLPAP KVDDAANWNA GLLYSKRLGP VGRAMIFQQT HTLFKSLSYL 540
541 KTESTDPFSN KRTRKAALWC VLTADAWLRC RIPFRAKPFV EEAKLFYGKI DWKDLKECVA 600
601 ALDEVEVSKQ PASNIPSS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
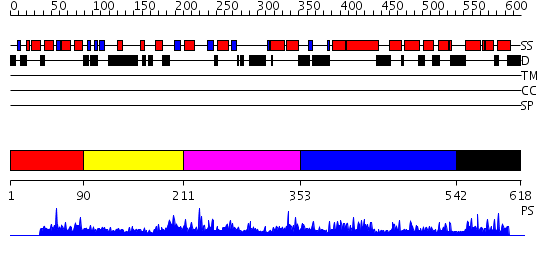
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..89] | 1.028994 | View MSA. Confident ab initio structure predictions are available. | |
| 2 | View Details | [90..210] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [211..352] | 1.036977 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [353..541] | 1.038997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [542..618] | 1.036989 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)