













| General Information: |
|
| Name(s) found: |
zds1 /
SPAC31F12.01
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 938 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell septum
[IDA cell division site [IDA cytoplasm [IDA cell cortex [IDA cytosol [IDA |
| Biological Process: |
fungal-type cell wall organization
[IMP ascospore formation [IGI regulation of protein localization [ISS] signal transduction [NAS] response to calcium ion [IMP regulation of catalytic activity [IC] |
| Molecular Function: |
protein phosphatase type 2A regulator activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSSVSNTL SIETKSDPKD PAFVASQEST ECNEHDTTQL SGSSSEPLEN NSSLTRSTDD 60
61 PSVEIRSKLV SPDNEANLLS DQNITISNEN NNENDTTEEA ETSSGNEAAD DEDSSSDAQS 120
121 SVPSFSEIHD GMSEEELDKE RKTLTHLRRI SLQGADDPEI PTDWSVAMSP PETEQDASTL 180
181 FWVPANLHPE LNPTGWKSFL DLQVKNLKSP TATDTSSSPL EHIRSLRRRK SLLSRQVKAD 240
241 DAVINYQDGS PIVEKAYLKR HRSLRLNELE HLESLARDPH RMVSLVDGMS NGSPEDSPLL 300
301 VSPNHFLQRS SRTTIRRTGA SIRTIHRGKT STLSGNRSHS ILQKPTDTSP LHKIEPISAD 360
361 ELVESDDSRT SALSNSQNPS DDVENQSDQA LEVLSLTNPP KIDNASADTT LHKETNKIDK 420
421 LYVSENKAES AVASESSLSE GTLALKAPAP ENKPEKSSTS KPPVPENKAE DSVVLKSSVP 480
481 EDKSENSIAS KPSATEGIPE NAIALQSSVP ENKAEDSVVL KSSVPEDKSE DSVPSKSSVL 540
541 EDKHENSVEI DKKADDSLPS NNKTEGYTPS VVREEKNYSE PNASPSVIPP RVPTPVPGRT 600
601 LSPKPTRIPT PIPSSLNVSL ESSKKPEIFH ERHIPTPETG PNKPSKNNIL KSTQVPVTPK 660
661 QKSSTANKGS TSSPSPPSSE SKKTKRSWGR LFVSGDSDKE HKEHKKDKQK KKNDQISSSS 720
721 KSASSFKKDR DKESIFGSLF GSKKKQTEIP PVSSSPPHND APPKAKPISA PSELPNTTSV 780
781 AEAKCQTVTD DEGTDQQSDE KSTEPKTFIP DKDYYWSRFP ICTERAIYRL SHIKLSNAHR 840
841 PLFQQVLLSN FMYSYLDLIS RISSNRPMNN VQQSTAKPIR KDINGQQRRS EFSAENVKNE 900
901 LENLSYQFGD QRKRNLNRKG STIHTVSQNI QKVSKNAK |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
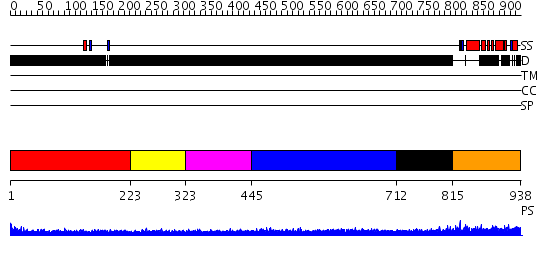
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..222] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [223..322] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [323..444] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [445..711] | 1.251996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [712..814] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [815..938] | 36.958607 | No description for PF08632.1 was found. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)