













| General Information: |
|
| Name(s) found: |
tps2 /
SPAC3G6.09c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 849 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA alpha,alpha-trehalose-phosphate synthase complex (UDP-forming) [ISS cytosol [IDA |
| Biological Process: |
trehalose biosynthetic process
[ISS dephosphorylation [IC |
| Molecular Function: |
magnesium ion binding
[IEA]
trehalose-phosphatase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSGAQGNTQ SAAYKRIVNV SRNFQFKAAL NKDTREWTFQ HRSSGTALYS AIASLGDPSS 60
61 PWDAVYVASP GPVQISEGND HTVETKIDEA AVVHRSSSIV ISPEDQEIFL KKATEYYRKK 120
121 NIEADIVPVW GKVQRVPQAK SSSATITPSV SNKLSTAYQS IKSEALKDGP MYADSVLWDV 180
181 LHYRIPTLDG YQQHNLWKNF TRWSQYFADN IVSNYRPGDL ILIHDYSLFL LPRLIRKQLS 240
241 DAPIVFFLHA PFCTSEVFRC LSKRAEILKG VLASNVIAMQ TDSYTRHFLS TCSNVLGLET 300
301 TSTHIDTSDG FRVVVFSSHV SIDVPRVYSR CNSKPVSLKI QQLKSLYPSD KKLIVGRDQL 360
361 TKACGVTHKL RAFRELLIHF PKWRGHVVLI QITSLPVGNN YSNEELKKTE NLVAQINSEF 420
421 GSLDYTPVIH FHQLLDPDEY YALLSVANIA CVSSVRDSMN TMALEYVACQ QKNKGSLILS 480
481 EFSGTAELLL SAYLVNPYDY SRFAETINYC LNMTEKERER RFSSLWKQAT SQSSQQWIYK 540
541 LINRAAYEVK ALESHMTTPL LTYNILIKPY RNAKRRLFLL DYDGTLIESA RNSIDAVPTD 600
601 RLLRTLKRLA SDSRNIVWIL SGRSQKFMEE WMGDISELGL SSEHGSAIRP PLAGSWSSCA 660
661 ENLDLSWKDT VRDFFQYYVE RTPGSYIEEK KHSISWCYQN ANTTYSKFQS LECQTNLEDM 720
721 LKHYDVEISP AKSFLEVHPN YLNKGSIVRR ILKRSGSVDF VFCAGDDKTD ENMFEVFLPT 780
781 AFSNSHLIDE IDSTEYSGSH HTDDNSDANK VENVKVATFR VHIGLTDKPT LSDYHLPAPR 840
841 DLGELLHNL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
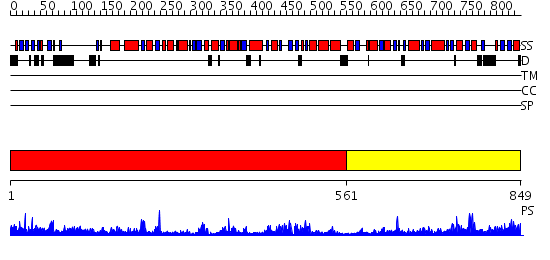
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..560] | 67.154902 | Trehalose-6-phosphate from E. coli bound with UDP-2-fluoro glucose. | |
| 2 | View Details | [561..849] | 27.522879 | Crystal structure of NYSGRC target T1436: A Hypothetical protein yidA. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)