













| General Information: |
|
| Name(s) found: |
SPBC6B1.05c
[Sanger Pombe]
|
| Description(s) found:
Found 21 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 649 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
autophagy
[IMP protein targeting to vacuole [ISS macroautophagy [IC] cellular response to nitrogen starvation [IMP |
| Molecular Function: |
small conjugating protein ligase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFVGKALQFQ SFHSSIDATF WHQLSNYKVE KQKLDASPLT IHGKFNTYSR GNISIVFGEA 60
61 PSNSNIKDCL AEGTLLNANT PQEFTNADVK KIREEIGEVL LNSIKNGVVS ERPNELLRFL 120
121 IFSYADIKAY KYHYWCLFPS FKETPHWIVK DLSPAESLIP SGPILSQIRE FLSTADYYQR 180
181 PFFLLIKSTL DEWTIAPLKE LSHCVDKSLQ FYLVAEDSVQ LAEYPSWPVR NILAFAFIKF 240
241 KLKVINLFLY RDGINSDTLS KSILIKVEAD KDMILEAPLS IVGWERNGKG VLGPRVVNLS 300
301 TVLDPFVLSE SASTLNLSLM RWRLVPQLDL DRIQNSKCLL LGAGTLGCGV ARNLLSWGVR 360
361 HVTFVDYSTV SYSNPVRQSL FTFEDCKRKL PKAECAAQRL KEIYPNMFST GYNISIPMLG 420
421 HPIYEAGIEK TMHDYETLEN LISTHDAIFL LTDTRESRWL PTVISTAMDK LLINSALGFD 480
481 SWLVMRHGSV LQKENRLGCY FCNDIFAPSN SLVDRTLDQT CTVTRSGCAN IATAIAVELF 540
541 VSLLQHPNGH AAPVLNEDQT VLGELPHQIR GFLHNFSLMK ISGMAYPQCS ACSECIINEW 600
601 NREKWMFVLR AINEPDYVEE LCGLREVQAL GEIAGTMEEW ISDKESVIL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
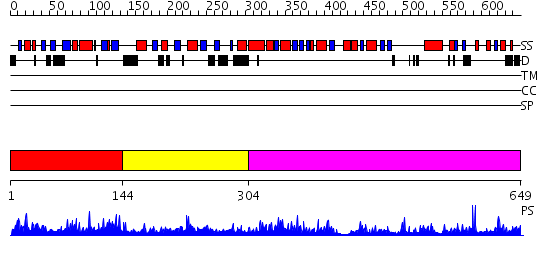
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] | 1.037976 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [144..303] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [304..649] | 65.154902 | Molybdenum cofactor biosynthesis protein MoeB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)