













| General Information: |
|
| Name(s) found: |
SPBC1709.13c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 547 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
ribosome biogenesis
[NAS]
protein amino acid methylation [ISS |
| Molecular Function: |
protein-lysine N-methyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKLLHEALQ NGCKLHKSVE FIQSRDDNAC FGSYIAVAQN DIAPDQLLIS CPFEYAITYN 60
61 KAKEELKKLN PNFESCNPHI TLCTFLALES LKGIQSKWYG YIEYLPKTFN TPLYFNENDN 120
121 AFLISTNAYS AAQERLHIWK HEYQEALSLH PSPTERFTFD LYIWSATVFS SRCFSSNLIY 180
181 KDSESTPILL PLIDSLNHKP KQPILWNSDF QDEKSVQLIS QELVAKGNQL FNNYGPKGNE 240
241 ELLMGYGFCL PDNPFDTVTL KVAIHPDLPH KDQKAAILEN DCQFQLSNLV FFLPKSPDKE 300
301 IFQKILQCLA VVTASSLELR KLTAHLLTGD LASYVPSLRG QIKSLEVLLM YIDSRADLLL 360
361 KSNPQVSPTS ERQVWAKIYR DSQINILQDS ITYVKNYMEE SLQKTYKPLP NLLQYLILNS 420
421 ISIFLLQHPL FAPLSHAIES LYGSTDAEAL VATDEQDILM ILICVYCLSI SEKLPFSISM 480
481 LVEGYPAVAN PEGVEVFEIL DEMFFQQFTN VFGESKHFNK ENVSWALQLV NDESLDFSGF 540
541 TFIIAHN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
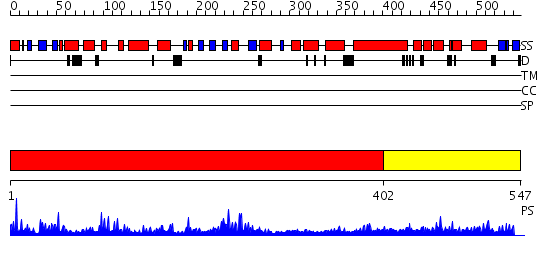
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..401] | 71.0 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain | |
| 2 | View Details | [402..547] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)