













| General Information: |
|
| Name(s) found: |
SPAC26H5.11
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 965 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA ascospore wall [ISS cytosol [IDA |
| Biological Process: |
ascospore wall assembly
[ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNEEDTDFAW LHNNSAEHLR FLSHRIFIGP IPSNFIHSTS GFFNKRFQNY TKRQICYNVA 60
61 SPNEPPIDFS FLMHKSTDEN PDTPSNLDSP STQNVGSTNN TRASQSLLRR SSSFFRRRHR 120
121 KNGTHASTDN NPFSESSTLQ PQTAERTSQQ AVRSAITETT NPSVSVQNSN STSTSSAAMI 180
181 IPHRDSQNSL EIAPLISPES QLSSLHPSSS RRHLISTPHV NRGTQFKRSS SCRNSRQPLL 240
241 SGVDKHLTSN FTDANLIIKQ SVVLARIEST FTVLPSDYND SAAQRVPRKT ISPWSQCLLV 300
301 ARQTDVENSI RLDFISKKLR KRLNGDVVHN LDVPHDKSYK SNYLFSVVLS PHQASWNIYN 360
361 SFDNSMVLWC PYGKNKTLIC LLNFQSSLLS FEWISIISRA LLFSPRPSLL ISVPAFHIHL 420
421 RLNFPCFKDT TRPHTNETFV TTDDITQLSR TSTLSLSTAS PRLVHDLVMK SWDISEDQFV 480
481 ESCLGVLEVN PEWSGIVKTW SKSHTLGLCW RMYDRLEWIN SFSSLKYVGL LAAKDLYQLE 540
541 LRPKLHYPNH VTFRDGSKMD EPTPVEGYLI RLTSSTGRKT RYGRMFHKEL YFAIFNNFLF 600
601 AIQPDSVLPL SMLSKSLNLD DKLPFLSNNE NDKYVYEFDP FKIANCRASG LNETIDSSVR 660
661 ESFLLLLQAE RKRELDMLTI ADSFLDLSRV ESVCPVEDVE ERNIFEITMT NGMKLVFQSY 720
721 CERTRNLWIN KITEVASYWK QRLFLDLQEY HDVRETNINI LHIEQSIEPD VACYLNHWEV 780
781 AGCVASTLIY HYCSMLGCRV IRMQGTLFKK KDALFEKCFA ILIPGQLVFF QDATRTKFGK 840
841 LCTKTHYRKR YSISLKNAFI YTGLSTMDEF ARGPNDPQPH ISRLPRCYED GWQSFDRDDM 900
901 LSFVIWSSGG VDYDLRPHHS VPDTAYGMAK DKLSKPKRFM FLARTRQERD VWAKRISKEI 960
961 NRGHS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
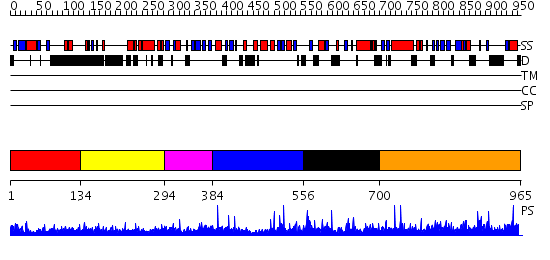
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 3.276997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [134..293] | 5.275996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [294..383] | 8.69897 | CREB-binding protein, CBP | |
| 4 | View Details | [384..555] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [556..699] | 1.81 | Solution structure of pleckstrin homology domain of human beta III spectrin. | |
| 6 | View Details | [700..965] | 1.42 | Crystal Structure of a N-terminal Fragment of SKAP-Hom Containing Both the Helical Dimerization Domain and the PH Domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 | No functions predicted. | |||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)