













| General Information: |
|
| Name(s) found: |
ero12 /
SPCC1450.14c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 571 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFMWMKIAKG LVGLLILYKN VCQVLCKSGS SVKSGSPWSI KNDGKSIIHD TLNSSYSDIE 60
61 NLNSRVSPLL FDLTEKSDYM RFYRLNLFNK ECRYNLDDNV ACGSSACNVL VTDEQDVPEV 120
121 WSSKSLGKLE GFMPELSRQI VETDRSVMEH VDKISQSCLL ERLDDEAHQY CYVDNELDSG 180
181 CVYVSLLENP ERFTGYSGPH STRIWEMIYN QCLPDSSAPT IDFPALFMQG PLAPPPKPQE 240
241 QLLKERMDAW TLEQRVFYRV LSGMHSSIST HLCHGYLNQR NGVWGPNLQC FQEKVLNYPE 300
301 RLENLYFAYA LMQRAIDKLY GHLDSLTFCH DSVLQDSEVR QKIAGLVSLI HNSPKMFDET 360
361 MLFAGDPSIS TALKEDFREH FKTVSALMDC VGCERCRLWG KIQTNGFGTA LKILFEVSNI 420
421 EDEVTNFDSR SFSLRLRRSE IVALINTFDR LSRSINFVDD FKQIYSEQHK PASFKRRVLR 480
481 RIKQLLFSVT PVALHPFLQK TSSILVDLYF DFKAEWDNVM LGFRYVFASY LRFPRLFKFV 540
541 LFSQESPFLN WTSHHLQRYI PKNWFPEVAS V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
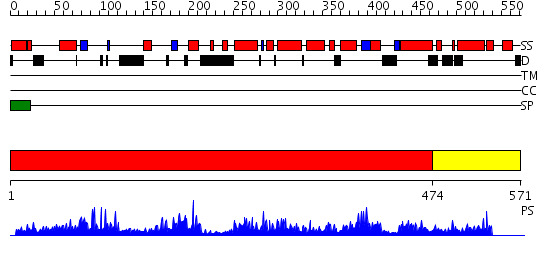
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..473] | 127.0 | Structure of Ero1p, Source of Disulfide Bonds for Oxidative Protein Folding in the Cell | |
| 2 | View Details | [474..571] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|