













| General Information: |
|
| Name(s) found: |
mnl1 /
SPAC23A1.04c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 787 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum lumen
[IEA]
membrane [IEA] cytosol [IDA endoplasmic reticulum [ISS |
| Biological Process: |
glycoprotein catabolic process
[ISS protein deglycosylation [ISS ER-associated protein catabolic process [ISS response to unfolded protein [IEA] |
| Molecular Function: |
calcium ion binding
[IEA]
[NOT]mannosyl-oligosaccharide 1,2-alpha-mannosidase activity [TAS carbohydrate binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSLHSIFCV CLILLCIFKE NSIVGAINSA RMVELRETSR RLFYHGYNNY MQFAFPNDEL 60
61 APLSCEGLGP DYENPNNIGV NDVRGDYLLT LVDVLDTLVV LGDREGFQDA VDKVIHHINF 120
121 ERDTKVQVFE ATIRILGGLL SSHIFASEEK YGFQIPLYKG ELLTLATELA ERLLPAFRTP 180
181 TGIPFARINL MKGVAYREVT ENCAAAASSL VLEFSMLTAL TGNNKFKASA ENAFFSVWKR 240
241 RSGIGLLGNS IDVLSGRWIY PVSGVGAGID SFYEYAFKSY IFLGDPRYLE VWQKSLESLR 300
301 HYTASLNEYY YQNVYSANGM VMSRWVDSLS AYFPGLLVLA GELELAKKMH LYYFSIYLKF 360
361 GQLPERYNLY TKSIELNGYP LRPEFAESTY YLYRATKDVF YLHVGELLLS NIENHLWTPC 420
421 GFAAVENLEK YTLSNRMESF FLSETLKYLF LLFDDDNPIH RSHHDFIFTT EGHLFPVTNQ 480
481 TRLSTSQRIY DGGEVCVAED YDRSKWPMLY SLIASRDDYD YVSHLVGIDN KAIPSYLIDP 540
541 SGVCKRPEYS DGFELLYGSA LVSPIKSVER VTNNVYISDI IGNKLKFVQK EGSNSLFLYT 600
601 AGAESINEND TVFLTDPDYE TFTRPDALYF DRTIAQLENP NSGQKTFGKF LQFDNDNSLP 660
661 SKVFKTVLLN NSMCQKPSDT LDKDTAYIAP LGNCSWVQQA KNTNKAGLLI LITDGEFINP 720
721 LENIHSQNSL FNWVKPYIPP TILLKNENNF VSKWANVSVR QSDFDAPSYF QGTPVSNLYL 780
781 CLKCINS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
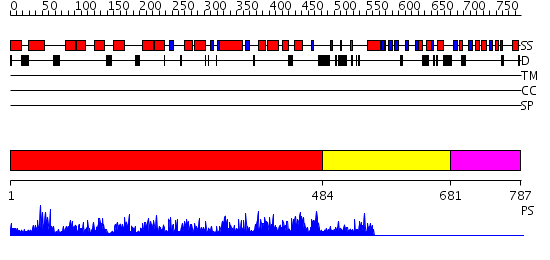
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..483] | 176.0 | Structure of mouse Golgi alpha-1,2-mannosidase IA reveals the molecular basis for substrate specificity among Class I enzymes (family 47 glycosidases) | |
| 2 | View Details | [484..680] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [681..787] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|