













| General Information: |
|
| Name(s) found: |
ucp3 /
SPBC21D10.05c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 601 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA cytosol [IDA actin cortical patch [ISS] |
| Biological Process: |
regulation of ARF GTPase activity
[IEA]
endocytosis [ISS] regulation of mitotic cell cycle [IGI |
| Molecular Function: |
zinc ion binding
[IEA]
ARF GTPase activator activity [ISS ubiquitin binding [ISS polyubiquitin binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGRRNETAI RELVQSVSGN NLCADCSTRG VQWASWNLGI FLCLRCATIH RKLGTHVSKV 60
61 KSISLDEWSN DQIEKMKHWG NINANRYWNP NPLSHPLPTN ALSDEHVMEK YIRDKYERKL 120
121 FLDENHSTNS KPPSLPPRTK SSSQSSPMAS TSTSKSRYAD SLSTLHDMGF SDDSVNTHAL 180
181 EETNGDVTRA IEKIVQHGSS KPQKPSTLTS TKSTIKLVSA RLKKRNKNLS VHFEDGTKPG 240
241 DAYEVTGDTR AASPTLNPFE QMMAMTNQGM SVSPGVETTS SPFFTAPVEP NQPLQPLRPS 300
301 MTGPVPSSMG SEATLNMPST YGIDSNLYTN SNSSSIVQNP LQPARTGPAA INYNYTTNYS 360
361 VSSPSVTNPF FDVGSSTQNA SLMGSTGYPS SANNVYYENS YQGVGTSMSD NYQLPDMSKL 420
421 SLNEQPAAPS NSNSQYMNTS MPTLDSTNMY GQSNQDPYSM SNGVYGSNYS AQPSTMQMQA 480
481 TGIAPPQPNM SMQMPMSMQS TGYQMPMENT WVDYNGMSQQ GNGMQPATMN YSNSMGYDTN 540
541 VPADNGYYQQ GYGNVMMPPD ASYTGTGSYV QPMNQPSGGM GAPADSSKAD SYIQRIMQGK 600
601 Q |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
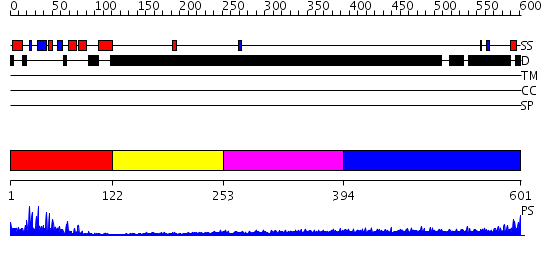
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..121] | 31.30103 | No description for 2iqjA was found. | |
| 2 | View Details | [122..252] | 2.12 | Solution Structure of RSGI RUH-016, a UBA Domain from mouse cDNA | |
| 3 | View Details | [253..393] | 1.266996 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [394..601] | 5.0 | Sec24 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)