













| General Information: |
|
| Name(s) found: |
SPAC688.14
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 461 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
protein amino acid methylation
[ISS |
| Molecular Function: |
protein-lysine N-methyltransferase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLKWAIEKDN YTTSEKIGLN DYRHVHESLG IGFIALEDIK EDEKLVSFKK DSVLCLTNSD 60
61 LAQLPEVQSL PSWAALLLVM ATENASPNSF WRPYLSIFPT KERITSPFYW DENKKDALLR 120
121 GTVLESNEDC NEITQLWIDR IEPIIKLYPN RFSQVSYEDF LRMSAVMLAY SFDIEKTKSP 180
181 ISNENEKSAA ETSIKEDKNG DAAKKNEGSA NQDDEKLHSQ SLVGNNCEVN SEDEFSDLES 240
241 EVDPDELEKA MCPISDMFNG DDELCNIRLY DINGTLTMIA TRDIKKGEQL WNTYGELDNS 300
301 ELFRKYGFTK KKGTPHDFVL IKKEHWLPEY IEKLGFEEVE ARLELLCREE LLYNLEGDFT 360
361 FSKADLTFKE ICLAFVLMEK EKELISVPSK SDIKPKHYRK LLKIIEKRIN MYPKISDPKN 420
421 FDEENAKTLI EGEIDILQNL SAKVKEALTK NRPKKKQKVD H |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
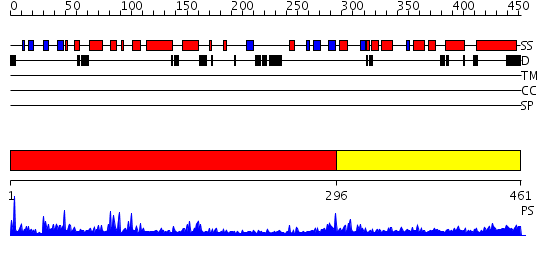
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..295] | 48.221849 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain | |
| 2 | View Details | [296..461] | 3.61 | RuBisCo LSMT C-terminal, substrate-binding domain; RuBisCo LSMT catalytic domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)