













| General Information: |
|
| Name(s) found: |
SPBC23E6.02
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1040 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [ISS chromatin remodeling complex [ISS |
| Biological Process: |
DNA repair
[ISS chromatin remodeling [ISS protein ubiquitination [ISS |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS DNA binding [IEA] ATP binding [IEA] zinc ion binding [ISS ATP-dependent DNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRNNTAFEQF TLNPCEQIPL DDKHNIGFNK NNTPDYSSSA SSDQLLKNDI NRHEMERRIA 60
61 FLNRKQALFN AFMHDSSSSL NTMESNIEKV NGLFPNDNSV IALKPNEEKL NSSLSVENND 120
121 STYTDATLIA PKIGLDRPNI NAITIDVDGH SLQNEISSST DKLSPSQSDA LFEQKQDSLF 180
181 WNDNAVIVVS DSESDDNNVR TKSSLNDHDK VNMKEKRNLE LAFMNSKRKK LELPSLPVLS 240
241 TAGPSYTNSL ALPPFHHHNN YKMFNTTHTL EDDKFLQGKG TSNNPISLSD EEDNEINFQN 300
301 KRYGSDSVIL PGGLLHDSKL PEPGKHLFHL QWYHDRFHNI EGFNLSDSNN QKVQDDQQQQ 360
361 LEELFKDLDE QLVNDPTIRE GTPAGLIPTL MEHQKEGLMW LKRLEESSKK GGILADDMGL 420
421 GKTVQALALL VTRPPESKSV KTTLIITPVS LLQQWHNEIL TKIAPSHRPT VYIHHGSSKK 480
481 HKIAEQLMSY DIVLTTYNVI AYEFKNKMAY DKSIEDNAPI KKFEHLPFFE AEWYRVILDE 540
541 AQTIKNRNTL AARGCCLLES TYRWCLSGTP MQNGVEEFYS LIKFLRIKPY SDWSSFSKDF 600
601 TIPLSSNINT SAPMKRFRGL LKAVLLRRTK NTKIDGKPIL TLPPKTAVKS ETDLSSSEME 660
661 FYNTLQSGAQ IQMRKYLQEG TITTHYGSLL VLLLRLRQAC CHPWLIVARE AAVDDNDSFQ 720
721 AKNRAIYNQI YPEAVNRLKL IETLQCSLCM DVVAELLIIV PCGHFLCREC LTHVITSSED 780
781 MAKQTSNENI SPKCSVCEEY IDTERLLSYA LFRRYSGMAP IVDADNKLRT ENISELLPKQ 840
841 YSNILENRQM GMKIFTDPKH WTTSTKIEKA LNAVKEIIKK QPTDKILIFS QFVSFLELFT 900
901 VPFRQEGIKY LMYTGGLSTA ERNQALINFE VDPNVRVLLI SLKAGNVGLN LTCANHVIIL 960
961 DPFWNPYIEE QAVDRAHRIG QDKPVNILRI VTNNTIEERV LALQDRKREL IDSALGEKGL1020
1021 REISRLNTKE LSFLFGMSSR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Liu X, et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
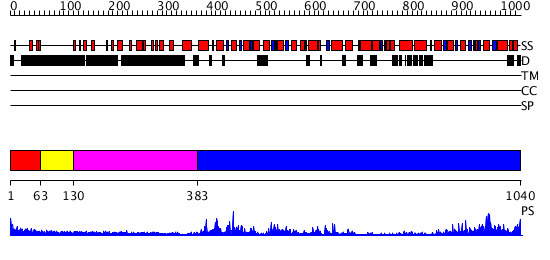
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..62] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [63..129] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [130..382] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [383..1040] | 39.69897 | Nucleotide excision repair enzyme UvrB |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)