













| General Information: |
|
| Name(s) found: |
SPBC1604.19c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 658 amino acids |
Gene Ontology: |
|
| Cellular Component: |
TRAPP complex
[ISS nucleus [IDA cell division site [IDA cytosol [IDA |
| Biological Process: |
ER to Golgi vesicle-mediated transport
[ISS autophagy [ISS peroxisome degradation [ISS intracellular protein transport [IC |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDIATNFITR SLSEIAPNAS QSSLHVTPSS VSNNAFGLKL RQLISDTYCP HIFIMASDDT 60
61 ENFMKRKGFD DFASFIRPFG DRIFQSSTNN SAISKETLDS RIFDNDHDSI RFVPMEAVQV 120
121 PRQWKRNKAW NTENGLENCA NSRFAPGGDI DSFRVLSQKL IEEWVQSGSN NYPENGFLNP 180
181 ILDYLKLLLS GNPVAEHETF SHPVGCLIVI TSHNTNPMAT VMRLFKEINN APFPNFISKE 240
241 ILHYYLFIHD EDNNDLSNSK QIFQQMRRSL GANSHFIRLR SNYISAKPLD TDRYDTSSIQ 300
301 SLKNAPRDSM ESESFCSSSD DAKPITFVTK NLRKFPIPEW RSSLEVQAES EQSCLPLYPL 360
361 LPVEEVEGMK KFVQTMLYDS IYPFMQRCVR AWEEDLTPQY GNLTTRLLFA SKKYWSRNHS 420
421 SHSQGNYDPL SLIYSSEKQE SIKRKMADFS FMLRDYKRAY EIYDEIRNTF SQDKAWNYLA 480
481 SCEEIQIICL LMQNRNISLK SQISYLNKWF DEMVYIYAVR LHSFYYTFRS VLVTSLLLSL 540
541 KPAFSIDFAA SWLAKILEPG PISLNPFETS FLNTTIAGMY SNKEHVGVTD GNRRRKAAFW 600
601 FAYSAGFWRD CGHCKMAEIC WNLANRVYSK SGWESLSEHM LDLKPTLSEN FRVKNTFH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
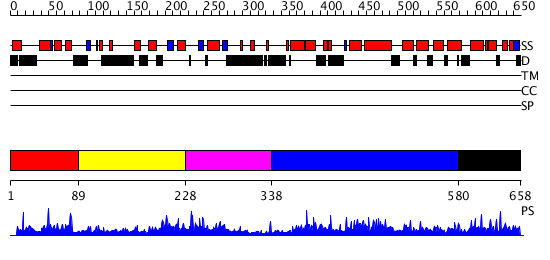
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [89..227] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [228..337] | N/A | Confident ab initio structure predictions are available. | |
| 4 | View Details | [338..579] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [580..658] | 1.016987 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)