













| General Information: |
|
| Name(s) found: |
SPAC26H5.03
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 512 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nuclear chromatin [IC] CAF-1 complex [IDA nuclear replication fork [IDA cytosol [IDA chromatin remodeling complex [IDA |
| Biological Process: |
ascospore formation
[IMP negative regulation of histone acetylation [IMP positive regulation of histone H3-K9 methylation [IMP heterochromatin maintenance [IMP positive regulation of chromatin silencing at silent mating-type cassette [IMP positive regulation of chromatin silencing at centromere [IMP protein localization in nucleus [IMP nucleosome assembly [IPI DNA replication-dependent nucleosome assembly [IC |
| Molecular Function: |
protein binding
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRAEVLQIRW HYDANDDHTP IYSVDFQKNS LNKFATCGGD SKIRIWQLIT SESSTKVEYL 60
61 STLSRHTQAV NVVRFNPEGN ILATAGDEGT IMLWVPTNTP ITTLADDAEE LALAKEYWKV 120
121 KIVCRSMGSE IYDLCWSVDS NFLIAGAMDN SLRLYDAHTG QLLTQKFDHS HYVQGVCWDP 180
181 LNQYIVSESS DRSICLYEIQ EEKKNPKKFQ LVLKSRICRI EYNVTKFELI SVTKPLNNDE 240
241 SSGISEPIET SNNNESPVSK HEALSSTANI VKDGSLERTE PPNSLNSKIS YSLYCNETLV 300
301 SFFRRPAFSP DGLLLVTPAG RLRPHGQPNF EVPYTAYIYT RGSITKQPVA CLNGFKKPVI 360
361 AVRFSPIHYE LNSFSNFSFT SVSFNLPYRM VFAVACQDAV YIYDTQTCKP FYRAVNLHYS 420
421 NLTDIAWNDD GNVLLMTSID GFCSVITFEP GELGVKSQHK ISLPEKRSAS PSSIDDSQDN 480
481 TAGGPATTTL IPRKVESSKV SKKRIAPTPV YP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
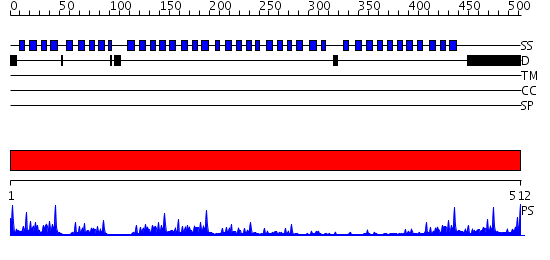
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..512] | 43.39794 | Actin interacting protein 1 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)