













| General Information: |
|
| Name(s) found: |
SPBP8B7.18c
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 551 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytosol
[IDA |
| Biological Process: |
phosphorylation
[IC thiamin biosynthetic process [IEA] |
| Molecular Function: |
ATP binding
[IEA]
phosphomethylpyrimidine kinase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPYNPLYESL SNQFDPSRIE TVLDPMGYTI KRRALPVSLT IAGSDCSGGA GIQADLKTMT 60
61 SLGVYGMSAI TCLVAENAGG VDSVEEMSPA FVESQIDCCI RDIPCHVVKT GMLGSPEIVK 120
121 AVARSAKKFN FSKLVVDPVM VATSGDSLVT KDIVSVLNEE LLPLTYLVTP NIPEAIVLAK 180
181 NQGLDISNIN SVSDMERCAA VIHKLGPKHV LLKGGHMPVN NLGLKSSDDE DLRVVDILYD 240
241 GNRFYHFSSS YLKKGEVHGT GCTLSSAIAS FLAWEHSLTE AVQFGIDYVH GAITHSPPIN 300
301 NCSTNILNHM TRLRIVPFAP GHFIEYILSH PQVVPAWKEY INHKFTNMLA KGTLPLPAFQ 360
361 DYLKQDYLYL VNFARAYSLK GYKENTFPNI LEAAQSVIHV IEEKELHVSM CSSYGVSLQD 420
421 LKSCEESPAC TAYSRYILDT GAAQDVAALD FVQAPCLIGY YVIAARLMKE PFRNPQGPYQ 480
481 KWVDNYFCED YLSAVRRGCR QIEEIVLKLS PERIQELIEI FIRATKFETL FWETPYYEYV 540
541 TKQNLEDKEF S |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
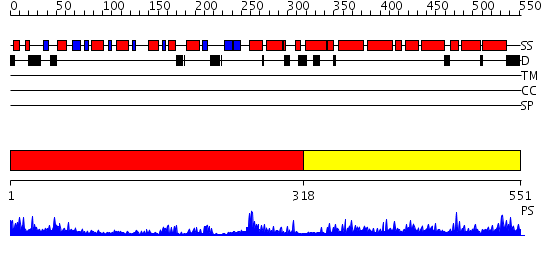
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..317] | 54.221849 | 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase (HMP-phosphate kinase, ThiD) | |
| 2 | View Details | [318..551] | 56.154902 | Structure of TenA from Bacillus subtilis |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)