













| General Information: |
|
| Name(s) found: |
SPAC24C9.11
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 775 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[ISS nucleolus [IDA |
| Biological Process: |
RNA metabolic process
[IEA]
translation [NAS] |
| Molecular Function: |
eukaryotic initiation factor 4E binding
[NAS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRPIKKSRSL KLPKSILEEI GESDSSARRG KRNHNLPHRE KRKFARISRG KNGYENRKIT 60
61 EEGDSKSSEL NDDYLDAHRK SSTKKKSSQN KQAKESKLLD PIAFQHKIAL EEDDREIAHL 120
121 EKMLGIKSKD KSAHSKIDKE FGWLLEDLDQ EIDDIGVPGT EDPSYGHSED SEVSDDTGDH 180
181 GSVDELESER EGNSGEEEEE FHGFESNSDE FHQPETKPIR MDPLKPAVPS LPNANVGSKY 240
241 VPPSLRKKLG GDKESEDALR LRRKLQGSLN KLSIANISSI IKEIEVLYME NSRHSVTSTI 300
301 TNLLLQTVMG RESMLDQLAI VYAALATALY RIVGNDFGAH LLQTLVERFL QLYKSKEKEP 360
361 LSSHKETSNL IVFFVELYNF QLVSCVLVYD LIRLFLRSLT ELNVEMLLKI VLNCGGQLRS 420
421 DDPTSLQDIV TEMNLLLASA DPSTISVRTK FMVESITNLK ENKKTKVANA SAQSKFEAVN 480
481 QLKKFLGSLG NRSLNAREPL RVTLEDIEQI ETKGRWWLVG ASWNNVPSGD NTLSTEALQD 540
541 KKKSEELTAH SKILQAAKKL RLNSTLRTSI FVALVGSEDY IDAWERVLKL HLKRNQLPEI 600
601 AYVILHCVGN EKLYNPFYGL VALKCCTLQH NLKKSFQFSL WDFFNELQPD DDSEEREISM 660
661 RRIVNLAKLY ASLVIEAAQP LTILKHVDFM AINAQMQTFL LVFFTDIILG VKDDLQLVKI 720
721 FENCKAEKNL SSKVDWFLKT YVRKNPLVDN SKKALFKSNL AMASAILQSI SKEEI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
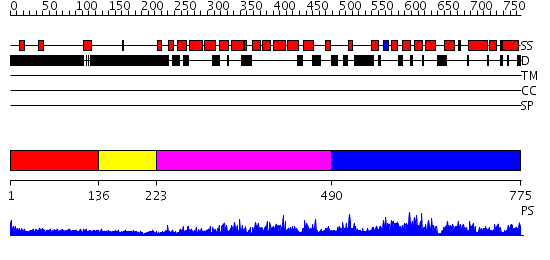
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..135] | 3.223996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [136..222] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [223..489] | 46.522879 | Eukaryotic initiation factor eIF4G | |
| 4 | View Details | [490..775] | 1.78 | CBP80, 80KDa nuclear cap-binding protein |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)