













| General Information: |
|
| Name(s) found: |
SPBC3H7.11
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 248 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA actin filament [ISS |
| Biological Process: |
actin cytoskeleton organization
[ISS |
| Molecular Function: |
actin filament binding
[ISS S-adenosylmethionine-dependent methyltransferase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSGISKLSDF WVEKYKKESK KSWDKFYKRN ETRFFKDRHW LDREFDCYFG LPDKLPLTIL 60
61 EVGCGVGNLV YPLLEVQPNL KIYCCDFSPR AIDFVKKHSC YNENRVFPFV NDITEDSLLE 120
121 VLGSACIDTL TAIFVLSAIP REKQLRSIKN LASVIKPGGH LVFRDYCDGD FAQEKFMTSG 180
181 DPSMIDEQTF VRQDGTLSLF FREEDIAEWM KSAGFGLVTL DRVNRTVDNR KRNLNMKRTF 240
241 LQGVWKKL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
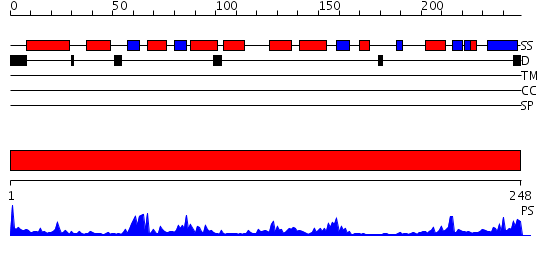
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..248] | 24.221849 | Crystal structure of Hma (MmaA4) from Mycobacterium tuberculosis, apo-form |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)