













| General Information: |
|
| Name(s) found: |
nta1 /
SPBC23E6.03c
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 286 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [ISS cytosol [IDA |
| Biological Process: |
proteasomal ubiquitin-dependent protein catabolic process
[IC asparagine catabolic process [ISS] N-terminal protein amino acid deamination [IC |
| Molecular Function: |
protein N-terminal asparagine amidohydrolase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKFGCVQFFP KLGKVNENIV HLRQLLDQHS EALQSVKLLV FPEMCLTGYN FKNSESIQPF 60
61 LENVTSNHCP SIQFAQEVSE QYRCYTIIGF PEFQNSNGIS TLYNSTALIS PKKELLNVYH 120
121 KHFLFETDKS WATEGKGFSF EPCIPELGPI SMAICMDINP YDFKAPFEKF EYANFILREL 180
181 EHQQMVSSNV SRPIICLSMA WLVSDDKVID ASLPDIKNLH YWTTRLSPLI NSNTDAIVLV 240
241 ANRWGKENDL NFSGTSCIME LSQGRAILHG VLKAAENGIV VGELEK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
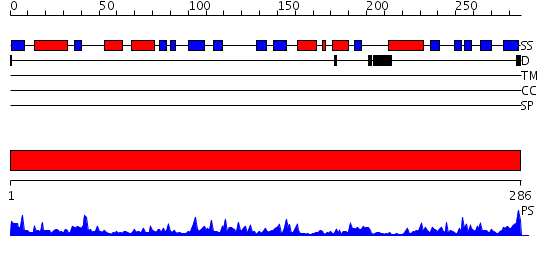
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..286] | 49.30103 | Crystal Structure of Hypothetical Protein PH0642 from Pyrococcus horikoshii |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)