













| General Information: |
|
| Name(s) found: |
SPCC825.01
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 822 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[NAS cytosol [IDA |
| Biological Process: |
ribosomal small subunit export from nucleus
[NAS]
ribosome biogenesis [NAS] |
| Molecular Function: |
ATPase activity
[NAS ATP binding [NAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARGKRSTQR DADLELESLQ SEIESESPQP VTKSKAKKNK KKLNKASAFN SDNDSNYDLK 60
61 PEDDEVDEEV VPVKKKPSKK SKKAKANAFE AFADEQSVEE EEEEDSEKPV RKNKKSSKKA 120
121 SPKNAFDALA DDMDDLSLDE EESESSEKSK KKKKKSKSKD DGSEALDDGD IESSEKDKKK 180
181 KKKSKENDDA PKKDRKTRKK EEKARKLASM LESENKDNDA NAAPLNKTDA FKDGLPSGRL 240
241 IFAYASGQKV APDGSNPADG ITVTGNLLSP PNSRDLQVEK LSVSAWGKLL IKDSELNLIN 300
301 GRRYGLIAPN GSGKSTLLHA IACGLIPTPS SLDFYLLDRE YIPNELTCVE AVLDINEQER 360
361 KHLEAMMEDL LDDPDKNAVE LDTIQTRLTD LETENSDHRV YKILRGLQFT DEMIAKRTNE 420
421 LSGGWRMRIA LARILFIKPT LMMLDEPTNH LDLEAVAWLE EYLTHEMEGH TLLITCHTQD 480
481 TLNEVCTDII HLYHQKLDYY SGNYDTFLKV RAERDVQLAK KARQQEKDMA KLQNKLNMTG 540
541 SEQQKKAKAK VKAMNKKLEK DKQSGKVLDE EIIQEKQLVI RFEDCGGGIP SPAIKFQDVS 600
601 FNYPGGPTIF SKLNFGLDLK SRVALVGPNG AGKTTLIKLI LEKVQPSTGS VVRHHGLRLA 660
661 LFNQHMGDQL DMRLSAVEWL RTKFGNKPEG EMRRIVGRYG LTGKSQVIPM GQLSDGQRRR 720
721 VLFAFLGMTQ PHILLLDEPT NALDIDTIDA LADALNNFDG GVVFITHDFR LIDQVAEEIW 780
781 IVQNGTVKEF DGEIRDYKMM LKQQIAKERE EERRIELEKQ KK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
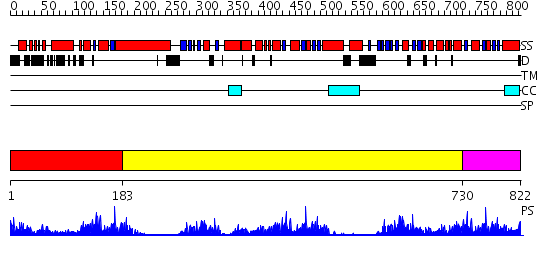
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..182] | 8.39794 | CRYSTAL STRUCTURE OF ATP-FREE RAD50 ABC-ATPASE | |
| 2 | View Details | [183..729] | 41.09691 | No description for 2iw3A was found. | |
| 3 | View Details | [730..822] | 59.221849 | RNase-L Inhibitor |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)