













| General Information: |
|
| Name(s) found: |
fbh1 /
SPBC336.01
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 878 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA ubiquitin ligase complex [ISS cytosol [IDA |
| Biological Process: |
DNA recombination
[IGI double-strand break repair via homologous recombination [IGI chromosome segregation [IMP replication fork processing [IMP regulation of DNA recombination [IGI |
| Molecular Function: |
magnesium ion binding
[IEA]
DNA binding [IEA] ATP binding [IEA] protein binding [IPI manganese ion binding [IEA] protein binding, bridging [ISS single-stranded DNA-dependent ATP-dependent DNA helicase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAQHLHSCK FYRLPLEIIP LICRFLSVQD IQSFIRVFPS FQTILDSSND LFWKKKNYEL 60
61 RIRRNRLLRG SYAAGVSSSA MNGFVNGTQI SSTPAEREYY SKSDEIKNIC GLPPGPMKVE 120
121 QIGHAIDHLV SETHTVDHLG SSIKASFFHI DDVPLEWISS ICMQVQSFFS PAAREAISSL 180
181 KSRTTTSNLL LSLFVGILFE DDLWYFFNTL YMLSSTSAIE FAYFLDSIFT VVRTDYEHYR 240
241 DPLSQTLITS CTRIHNIVAV IESPYDEPNT KGLTSEQKMI VECQLNPGEV LKVKAFAGTG 300
301 KTKALLEFAK SRPKDKILYV AFNKAAKEDA ELRFPFNVKC STMHGLAYGA ILAQADLPQA 360
361 KLERQLSNST IASLLSLQVA FPKANRKNNP GTPSASLVAS HIMFTLNRFM HSTDWQLGFR 420
421 HISKRSLEVT KLSKEKLLAY TKKLWSLIVN FEYTHAPLIP DAYMKLLHLY EFPNIFSKYD 480
481 YILFDEAQDF TPCMVDLIYR QKHARIVIVG DAHQCIYGFR GANACAFNEN LYPSTKQLCL 540
541 TKSFRFGNSV AKYANFLLSL KGENVKLKGV QNDHAYWSSA SNPNNVSGAF RFFPHTIIFR 600
601 TNKELILQSI RLSVSLPKEI PIAILGSMRK KAFQLLRSGS ELAHGQRPSH PKLKDFSSWG 660
661 EFEVHVKNSA EEDAELALVY DMADELFSES FLSRLDNCEK RLMDSKDDGD NGIILATAHQ 720
721 SKGLEWDNVQ LGNDFRPKFD SVSFSRIGSS RYLQEEINIL YVALTRAKKR LILNDTITKL 780
781 YALECGLVRF AGGILTEDQL QPGKVALFVD WQIDKFSFFY ETPAEGYNLL VEANEKSVWD 840
841 IFFGVLSGAW QNYIANTSER LKRSMLFIEN QLFAVHDQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
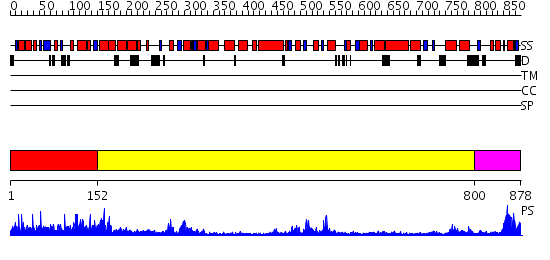
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [152..799] | 36.522879 | No description for 2gjkA was found. | |
| 3 | View Details | [800..878] | 96.522879 | STRUCTURE OF DNA HELICASE |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.53 |
Source: Reynolds et al. (2008)