













| General Information: |
|
| Name(s) found: |
rsv2 /
SPBC1105.14
[Sanger Pombe]
|
| Description(s) found:
Found 18 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 637 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA |
| Biological Process: |
cellular response to stress
[IEP positive regulation of transcription from RNA polymerase II promoter, meiotic [IEP gene-specific transcription from RNA polymerase II promoter [RCA positive regulation of transcription, meiotic [IEP |
| Molecular Function: |
transcription factor activity
[ISS zinc ion binding [IEA] specific RNA polymerase II transcription factor activity [RCA transcription activator activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTTNNTYAR SISSNGNDNF PTPDWQFNGS QLQQNRKSKM NGRSVTNVFP NAFSDAEFEQ 60
61 ISMPELNLKR FNPTTLEENN SDLSDMSSWD YMMNVPIRVY NDADTMDPFS LDTIPDSSMD 120
121 MPFDPLASDY GNAANFPSVP SSLGSNHQFI TTPPVNGSNE PTSAQTNHII TANSSPSGNA 180
181 GSNASASMSV PPPLTPSAST INDQPFSNSF DLPSQVIADG TGAISDINGN PFPMNSPPLD 240
241 MEPLPSISMD ASDSVSEQLV KDASLPSGPF STDYLENGSD LKRSLGHNQK SDRVSKDVSP 300
301 QHQANPSTLN NPLKTQNFDS SKNLYTDNKD SSLVSPTGLQ SRMEQNPEVR AHPMKDSATS 360
361 TALRRSHALG AAADSLLPQE NSAQIYDGKD VSMVNDNMHS DVRQDSFNKE SIKQRIPSLS 420
421 PPITRSYNAK HRPSLVLGTS VNPHSLSPSQ PPVVVPSNTT ISSSPPLTSP VKTSANIPNL 480
481 LPTSELDSSN APHSQSAATH DLNDVKSYYN TRSSHSVVPN PTNQKVSITG AAADGPNGSA 540
541 PVDTTPTNSS TTATGAQRKR RKFKFGKQIG PVRCTLQNRV TGEICNTVFS RTYDLIRHQD 600
601 TIHAKTRPVF RCEICGDQRH FSRHDALVRH LRVKHGR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
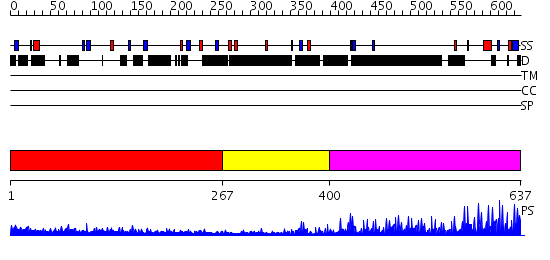
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..266] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [267..399] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [400..637] | 37.0 | No description for 2i13A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)