













| General Information: |
|
| Name(s) found: |
zfs1 /
SPBC1718.07c
[Sanger Pombe]
|
| Description(s) found:
Found 20 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 404 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytosol [IDA |
| Biological Process: |
mitotic cell cycle spindle assembly checkpoint
[IGI negative regulation of mitosis [IMP mitotic cell cycle checkpoint [IMP negative regulation of septation initiation signaling [IGI mRNA catabolic process [IMP pheromone-dependent signal transduction involved in conjugation with cellular fusion [IMP regulation of conjugation with cellular fusion [IGI |
| Molecular Function: |
AU-rich element binding
[IDA zinc ion binding [IEA] protein binding [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVYSPMSRPQ VPLAFRQWPP YNYKSDPLVN SKLSQSTTSV NAGPSLISPS FLDSYANSSS 60
61 LLHKPSTNLG SLKTSSLLAS DEVFPSSVMP QLRPLDSSLS VSPEMDGWPW HHNSSSNPQG 120
121 YAWTPSLLSS NATSYLHSGS SPHGNTSNHP SPISSLESLP SRSSTGSGSL DFSGLANLHD 180
181 DSKSLAMSLN MAGVPVSVDE NSQTSFPFVH GQPESTMSRK PKLCVQSKSM TNIRNSVAKP 240
241 SLVRQSHSAG VIPIKPTASN ASIRNAPSNL SKQFSPSGNS PLTEASKPFV PQPSAAGDFR 300
301 QAKGSASHPH GSGSSNGVAP NGKRALYKTE PCKNWQISGT CRYGSKCQFA HGNQELKEPP 360
361 RHPKYKSERC RSFMMYGYCP YGLRCCFLHD ESNAQKSATI KQSP |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
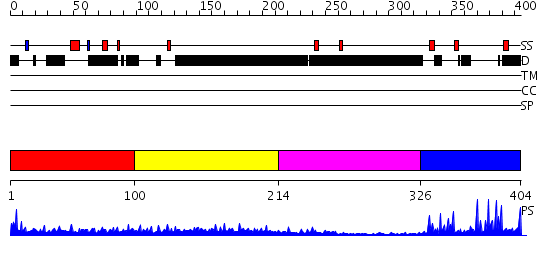
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [100..213] | 8.117995 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [214..325] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [326..404] | 20.154902 | Structural Basis for Recognition of the mRNA Class II AU-Rich Element by the Tandem Zinc Finger Domain of TIS11d |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)