













| General Information: |
|
| Name(s) found: |
SPAC607.06c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 612 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cell division site [IDA cytosol [IDA |
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
zinc ion binding
[IEA]
metallopeptidase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSQIILDNIS QNETVYNRFV IIHGRVGPRA SYCPQTINVK HHENSFPEQT WVVTDLYFKA 60
61 IIHVVPGDNT ILFTTDDGGK LELQVYYQLL VDNPFYCIAF IVGKDSDLSF DAPHGAKNDI 120
121 DEGIRKLRCA AYLWQAFTAE CMYRNGYGRR SFRIEESVQP DTMSCLSPWG TERLTATINI 180
181 LRSDKTAEEI RSVPPDQLFH IAGDAVDKLH LPEPWHYMCM FLDTRYDPST QKIRGHVALG 240
241 GGTDMHKLGV FGSHSLHSFP SALEYVVPVF SDARRIPSYL ANDANESSTI WECANIGIGA 300
301 MLHELGHTLG CPHQPDGIML RSYPIFNRSF TTREFECVRT GSKGLAPVLA KDECSWHHLD 360
361 MLRFFYHPCF KLPSDPTYPS DIETNYFVKG ETITFVNSTG IFLIEIEYNG QTKGWKEFNP 420
421 PVGEASLTDA EIRSLSNASS NQDYRVRVLS RNFKCIDLNN VPEIIKNAKV ESSFGTVYRS 480
481 ERYGLRGCNG KELSNILIAP EKFKPTKIRV HCGLALDGIE VFFGDESVLL GNRGGSPHDF 540
541 EIQGSQIVGF QIRCGAWVDG ISIVLENGKT SPFYGNANGG GLKSYLVPKG FQLVGFYGTL 600
601 GPFMDSIGFF IK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
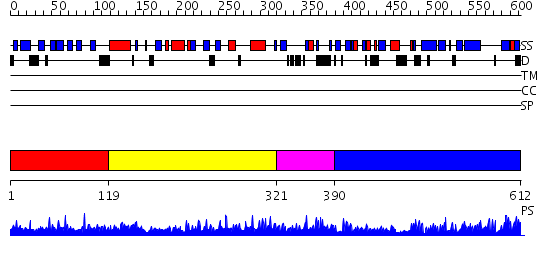
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [119..320] | 1.02 | Macrophage elastase (MMP-12) | |
| 3 | View Details | [321..389] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [390..612] | 34.154902 | Crystal structure of the Parkia platycephala seed lectin |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)