













| General Information: |
|
| Name(s) found: |
SPAC4F10.09c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 860 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Noc1p-Noc2p complex [ISS nucleolus [IDA |
| Biological Process: |
ribosome biogenesis
[ISS |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATLYPKPKR GQKIIFNEEG EGTIDTKPQG VMFDPGVPWY NIPLPELHED KKFVNVDHVS 60
61 ELETRGLLLL KEDSERFSET LGHGTADKRM LQTLISSGTT SDRISALTLL VQESPIHAVK 120
121 ALETLLSICS KKSRNEATQA ITTLKDLFIG GLLPDRKLKY MKQQSCLGSK NVTDKHLMVW 180
181 AFESFLKSFY FKYIQIIEAL SFDALLFVKS QMVSTIYDLL KAKPEQEQNL LKLLINKLGD 240
241 KENKIASKAS YSILQLEASH PAMKLVITKE IERFIFAPST SRTSCYYTLI TLNQTVLTHK 300
301 QVDVANLLIE IYFVFFTKLL FALEKEEVAD APTLEKKSLQ SDSKNKKSQK RKKDEDLRKE 360
361 AEENVNSRVI SAVLTGVNRA YPFAEVNSEK FDKHMNTLFA ITHTASFNTS VQVLMLIFQA 420
421 SASRDFISDR YYKSLYESLL DPRLTTSSKQ SLYLNLLYKS LIIDNNIPRV RAFIKRMVQV 480
481 SAWQQPPLVT GLFHVMHQLV IATTALRSMF TNAEIHDFDG DEEEVFKDVE EDDVSEDQKV 540
541 DSDKDGKLSD KQSHSAYVVG NVSVSTKKEH LSYDGRKRDP QYSNADGSCL WEIHPFLNHF 600
601 HPTVSLLAKS LVYGEKILGK PNLSLHTLNH FLDKFAYRNP KKSAAARGHS IMQPLAGGLS 660
661 KGYVPGSTYS GVPMNSEQFT SKKQEEIPVD ELFFYRFFND KYIKGKQARK TKVDRDEEGE 720
721 IDEDEVWKAL VDSKPQLEMD EEESDFDSEE MDKAMTDMGS DSEQSADEND NESMASEEKP 780
781 MFSDEENLSE IAHSEDEFDD TVDFFEDEND LLPFNETDDE EEIQTVDHSE THSHKKKKRK 840
841 AIKDLPVFAD AESYAHLLEN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
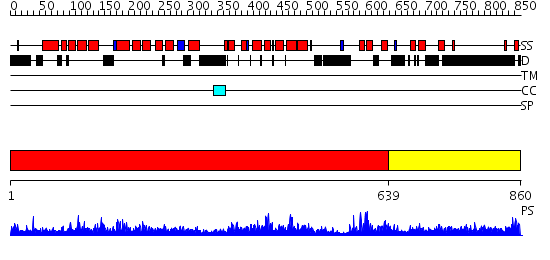
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..638] | 1.57 | Adaptin beta subunit N-terminal fragment | |
| 2 | View Details | [639..860] | 10.060996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)