













| General Information: |
|
| Name(s) found: |
SPAC4A8.10
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 723 amino acids |
Gene Ontology: |
|
| Cellular Component: |
lipid particle
[ISS cytosol [IDA |
| Biological Process: |
lipid metabolic process
[ISS cellular lipid catabolic process [ISS] |
| Molecular Function: |
lipase activity
[NAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTISSGENIS KQISENNSST NPKHANSESS PLLSSDFSSS SKDYHKVGAF TDNTNAINIP 60
61 EQTRGVSHTS SSPSVGTSFS SINLREVKNP LAGSDSTAPF INSTNSVLWI RVRNREARFR 120
121 QAAYLQGPFT LCVSVWNDQF FDEKNSLLNF DPQVQPSTSF WVCVPYSCFS NQQPIYIEIA 180
181 SQAIFKDRSI NFELAISQSQ HSIRAMSAAD IDTLHAFPAL HVEHQTPENL WRLPYSSFHS 240
241 KNSHLVVLTH GMHSNVGADM EYLKEKLIES SKSVKELVVV RGFTGNYCQT EKGVRWLGKR 300
301 LGEWLLDITG WGSASFPRYS HISVVAHSLG GLVQTYAVGY VHAKTHGAFF QAIHPVFFVT 360
361 LATPWLGVAG EHPSYIGKAL SYGIIGKTGQ DLSLTPLNHS IESRPFLVLM SDPSTPFFQA 420
421 VSFFEKRILF ANTTNDYIVP FGTSAMEVSS LGKVEEAEGS DKVMPTHMEN GISPTLKENE 480
481 QTVQSVGDNA KKIHASSEES GSSFSKALKS FVGLFSYSAS KTTDTEIPLV KNEDENARKP 540
541 TEPNCLGSDE LDVSNSSNQF FCSAPKLDPT STFSGVAQRV VNTFTNLFIP AVPTDSYFFK 600
601 YHLHKVVVSD NVHDPAASFS TFDGITELNN MNNGFLSNEI TIAKNWHRLA WRKVAVRLDG 660
661 DAHNTMIVRR RFPNAYGWTV IKHLTEILFE QNTSTAYMNP ISFDETTTAA WLSEIYEDNS 720
721 ISV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
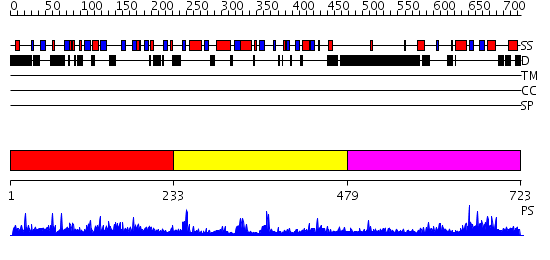
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..232] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [233..478] | 26.69897 | The crystal structure of palmitoyl protein thioesterase-2 reveals the basis for divergent substrate specificities of the two lysosomal thioesterases (PPT1 and PPT2) | |
| 3 | View Details | [479..723] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 |
|
||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)