













| General Information: |
|
| Name(s) found: |
mtr4 /
SPAC6F12.16c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 1117 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [ISS TRAMP complex [IDA |
| Biological Process: |
mRNA export from nucleus
[ISS tRNA catabolic process [ISS rRNA processing [ISS ncRNA polyadenylation involved in polyadenylation-dependent ncRNA catabolic process [ISS |
| Molecular Function: |
ATP binding
[IEA]
ATP-dependent RNA helicase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFGGELDDAF GVFEGKVPKS LKEESKNSQN SQNSQKIKRT LTDKNASNQE QGTKKLESSV 60
61 GEQESATKRA KIENLKDNQD LIPNNDVNGI HINNSAVADT KHKPKIGDIA ADDISNEVSI 120
121 KNEGDTIPEA TVADSFEQEA SLQVAGKVGM TEAKSSTEEV VELRHQVRHQ VSIPPNYDYV 180
181 PISKHKSPIP PARTYPFTLD PFQAVSIACI ERQESVLVSA HTSAGKTVVA EYAVAQSLRD 240
241 KQRVIYTSPI KALSNQKYRE LLAEFGDVGL MTGDVTINPD ATCLVMTTEI LRSMLYRGSE 300
301 VMREVAWVIF DEIHYMRDKE RGVVWEETII LLPDKSHFVF LSATIPNAMQ FAEWITKIHR 360
361 QPCHVVYTDF RPTPLQHYLF PSGSDGIHLV VDEKSNFREE NFQRAMSALM EKQGDDPAAM 420
421 ATKGNAKKGK TGKGGVKGPS DIYKIVKMIM VKNYNPVIVF SFSKRECEAL ALQMSKLDMN 480
481 DQTERDLVTT IFNNAVNQLS EKDRELPQIE HILPLLRRGI GIHHSGLLPI LKEVIEILFQ 540
541 EGLLKVLFAT ETFSIGLNMP AKTVVFTNVR KFDGKTFRWI SGGEYIQMSG RAGRRGLDDR 600
601 GIVILMIDEK MDPPVAKSML KGEADRLDSA FHLSYNMILN LLRVEGISPE FMLERCFFQF 660
661 QNSLEVPKLE AKLEESQQHY DSFTILDERP LEEYHTLKTQ LERYRTDVRT VVNHPNFCLS 720
721 FLQGGRLVRV KVGNEDFDWG VVVNVSKRPL PKGQSNEYLP QESYIVHTLV MVASDTGPLR 780
781 IRSGHLPEVH PPAAEDKGKF EVVPFLLSSL DGIAHIRVFL PNDLKSQGQK LTVGKALSEV 840
841 KRRFPEGITL LDPVENMNIK EPTFIKLMKK VNILESRLLS NPLHNFSELE EKYAEYLRKL 900
901 ALLEEVKDLK KKLSKARSIM QLDELNSRKR VLRRLGFTTS DDVIEVKGRV ACEISSGDGL 960
961 LLTELIFNGM FNDLTPEQCA ALLSCLVFQE KSEVENQRMK EELAGPLKIL QEMARRIAKV1020
1021 SKESKQELNE EEYVNSFKPS LMEVVYAWAH GASFAQICKM TDVYEGSLIR MFRRLEELIR1080
1081 QMVDAAKVIG NTSLQQKMED TIACIHRDIV FSASLYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
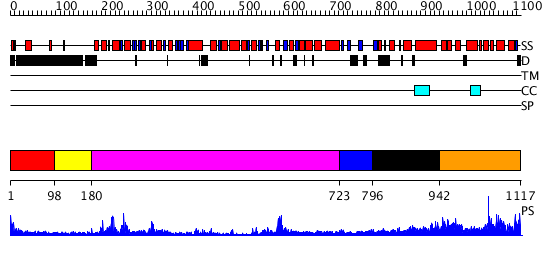
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [98..179] | 20.69897 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 3 | View Details | [180..722] | 45.30103 | Structure of the RecQ Catalytic Core | |
| 4 | View Details | [723..795] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [796..941] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [942..1117] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)