













| General Information: |
|
| Name(s) found: |
SPAC17G8.02
[Sanger Pombe]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 330 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [ISS |
| Biological Process: |
pyrimidine nucleoside catabolic process
[ISS NAD biosynthesis via nicotinamide riboside salvage pathway [ISS] nicotinate nucleotide salvage [ISS] pyrimidine salvage [ISS |
| Molecular Function: |
ribosylpyrimidine nucleosidase activity
[ISS]
nicotinic acid riboside hydrolase activity [ISS] nicotinamide riboside hydrolase activity [ISS] uridine nucleosidase activity [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTNTIDSFQK GSALENYNIW IDCDPGHDDV VALTLAACAG HCKILGVSTV HGNTTLEFTT 60
61 KNALAVMELL NQDVDVHAGA AKPLMRESAF ATHIHGTNGL AGISLLPDYP KKKATPDAVF 120
121 AMYTTISNYP EPVTLVATGP LTNIALLLAT YPSVTDNIER FIFMGGSTGI GNITSQAEFN 180
181 VYADPEAARL VLETKSLIGK LFMVPLDVTH KVLLDANIIQ LLRQHSNPFS STLVELMTVF 240
241 QQTYENVYGI RNGVPVHDVC AVALALWPSL WTSRSMYVTV SLDSLTLGRT VCDVWSQQNQ 300
301 YPANVHVVLE ADVSLFWETF IGVIDRLNYL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
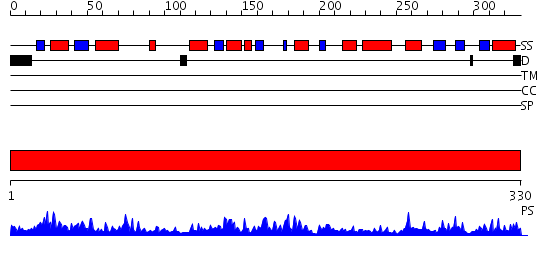
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..330] | 99.0 | Crystal structure of a the E. coli pyrimidine nucleoside hydrolase YbeK with bound ribose |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)