













| General Information: |
|
| Name(s) found: |
SPAC4G9.19
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 270 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
ER-associated protein catabolic process
[ISS protein folding [RCA |
| Molecular Function: |
Hsp70 protein binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKIFFRLNL VRNSLWRRAA SFTSYSELKS LTPYEILELP RTCTANDIKR KYIELVKKHH 60
61 PDKMKNASQL APTESPPEIN KHNEEYFRLL LAANALLSDK RRREEYDRFG IHWNQPSHPT 120
121 HPSPQNWSQA RYPTYSRSRR SAGMGSWEEY YYNSYDYMND QNASNKNRKF DDEGMLVFAG 180
181 ILSILVIINI YSNYRNGKFY REARSAAIGR AEDNFDYYST GMADLSKDDR ISRFLILREQ 240
241 SKQIPTQQKP SSLPPPERAL PAPTMPTPSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
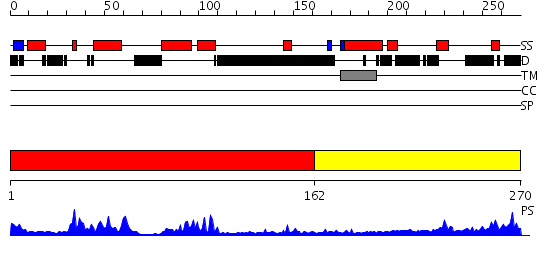
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 22.30103 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [162..270] | 2.251996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 |
|
||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.32 |
Source: Reynolds et al. (2008)