













| General Information: |
|
| Name(s) found: |
ppc89 /
SPAC4H3.11c
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 783 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA spindle pole body [IDA |
| Biological Process: |
mitotic spindle organization
[IMP chromosome segregation [IMP septation initiation signaling [IMP cellular protein localization [IGI |
| Molecular Function: |
protein binding
[IPI structural molecule activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPSQFNDNFV ATDDDVGTMA RVGMALDREL NGDSFQDNLN FQPRSGKRED FEPFFQDTPN 60
61 TKSLSKHFHD FTMNASLETL PSVEKPRGRN MNAFEETSWN RFRKNGSLFP LSSPPISEPD 120
121 LRPQALNETP DYRNRFLGAF KKQGVLDDHG NLKLDASPSF LKKPAEYTPL ANRQNQNLAF 180
181 DSPTEALPPK PTTPWRRNGF RSKTTPNLNS GKETPSSYKA SARLMEQLGL NHSEPSVDFN 240
241 NQTSYRLPNL TNLSSLIRDD TIDENGNAKE HDRLPELNTI PVASTDEQLF NAHQLLEKKF 300
301 EILKRERNEC NAKIDELQDK LELLTDAYNR EKRRARSLEE RMSKEMLTKL GESNVDDGMA 360
361 ASRYDTVKRE KERLSEHLKS LQEQYEHIQS VYKNVLLDRE SYIMRLGNKI SENNELLNEN 420
421 RVLKEKLQTY LDKKESNVTS KIKSTAENSS KPLSMNEADE RKDGLNNLLF ENKSGANTKE 480
481 MSNGTETAKE NCSPQQDSTS PTSGYQDLVK ELAKEIEMRK SLELKLKLSQ SNKAGPVKHR 540
541 KRRPKSKRRI TGKVVFDSPN VASGVESDEG SEEISLDSEY SDILSDDGDF EKEKQATLPR 600
601 RRSSSSMKGN KLAEDSYLNE AGFDWNQGTF HNGSEFGTTG VPDEPNEEEL PKHVLKQVEH 660
661 IINESAAHGV GKCNACHARQ EDLIRGEQKV SHSNCLYADQ TLRPSQPPSE ALKTVVNQLT 720
721 NELMELKKRY EKLSDRYNSL TPGYHKHKRQ EIKNKLIKLI ECMESKSDQI YLLYDVNVGK 780
781 DFS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
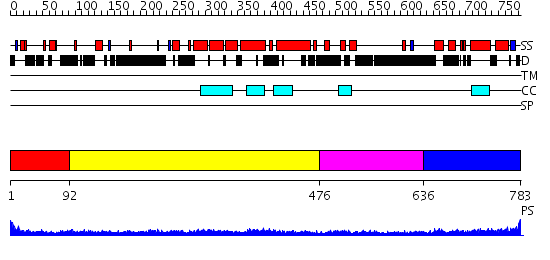
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [92..475] | 17.69897 | Heavy meromyosin subfragment | |
| 3 | View Details | [476..635] | 6.69897 | Tropomyosin | |
| 4 | View Details | [636..783] | 1.468994 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)