













| General Information: |
|
| Name(s) found: |
SPAC4H3.01
[Sanger Pombe]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 392 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cell division site
[IDA cell tip [IDA cytosol [IDA |
| Biological Process: |
protein folding
[RCA |
| Molecular Function: |
Hsp70 protein binding
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATPVDTEYY DLLGISTDAT AVDIKKAYRK LAVKYHPDKN PDDPQGASEK FQKISEAYQV 60
61 LGDEKLRSQY DQFGKEKAVP EQGFTDAYDF FTNLFGGAPF REWVGELSFV KEMFREEDSA 120
121 VEQGQMNDKQ QLLLESSEPT PTIKQQFNDR KKNAQIRERE ALAKREQEMI EDRRQRIKEV 180
181 TENLEKRLDD WIAKATTEEG LNALREKYTQ EANTLRIESF GVEILHAIGE VYTQKGRTVL 240
241 KSSKFGIGGF WSRMKEKGKI ARATWDTVSA AMDAKLSIDQ MQKLEDKGED QASAEERAKL 300
301 ELDITGKILR ASWCGARYDI QGVLREACSN LLKKRVPTEL RLKRAHALLE IGTIFSNVEA 360
361 DPDDPNRIFE NLILENKKKR KKGSEAKPPK AT |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
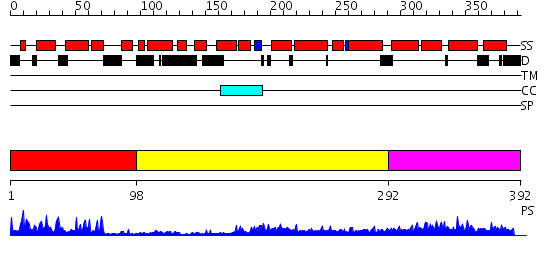
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..97] | 19.69897 | NMR STRUCTURE OF THE J-DOMAIN (RESIDUES 2-76) IN THE ESCHERICHIA COLI N-TERMINAL FRAGMENT (RESIDUES 2-108) OF THE MOLECULAR CHAPERONE DNAJ, 20 STRUCTURES | |
| 2 | View Details | [98..291] | 12.045757 | The crystal structure of Hsp40 Ydj1 | |
| 3 | View Details | [292..392] | 4.097992 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)